Introduction
Diabetes mellitus (DM) is a worldwide health issue
that affects 463 million individuals globally (1). Diabetic peripheral neuropathic pain
(DPNP) is a frequent and devastating complication of DM, affecting
25–30% of patients with DM (2,3).
DPNP is characterized by spontaneous pain, mechanical allodynia,
paresthesia and numbness in the terminal phase of DM and can lead
to lower-limb amputation (4).
However, the presentation of DPNP in patients is inconsistent. The
clinical presentation of DPNP largely depends on the affected
primary sensory fibers, although the symptoms of patients with
large-fiber involvement typically manifest as numbness and loss of
sensation, whereas those with small-fiber involvement more often
present with pain and dysesthesia (5). The discovery of effective
therapeutic-targets for DPNP is critical because of current lack of
effective therapies and most current guidelines recommend the use
of duloxetine, amitriptyline, gabapentin or pregabalin as initial
analgesic for the treatment of DPNP, which are effective for
symptomatic relief, but not for disease modification (6). Deeply understanding the pathogenesis
of DPNP might help explore mechanism-based approach to deal with
this difficult problem. Diabetes is a chronic metabolic disease and
alterations in the internal environment might revise gene
expression through epigenetic modifications in the occurrence of
DPNP.
Epigenetics is the biological process of long-term
heritable changes in gene expression that are influenced by the
environment, which is able to inhibit or stimulate protein
expression at the transcriptional level (7). Epigenetic modifications that are
involved in the regulation of chromatin structure and gene
expression have been reported to serve a role in the progression of
DPNP (7). One of the mechanisms
considered to contribute to the development of DPNP may affect
primary sensory neurons, causing altered genes expression, impaired
trafficking and the abnormal functioning of receptors and ion
channels (3,5). In addition, neuronal and glial cells,
such as microglia and astrocytes, are also involved in DPNP through
the production of powerful neuromodulators including
pro-inflammatory cytokines and chemokines, which increase neuronal
excitability. Abnormal expression levels of pro-inflammatory
cytokines and chemokines, neuronal receptors or ion channels have
been linked to epigenetic modifications involved in DPNP. These
epigenetic modifications include DNA methylation, histone
methylation and acetylation, non-coding RNA (ncRNA) and RNA
modifications. A number of previous reviews have discussed the
importance of epigenetics in the pathogenesis of neuropathic pain
(NP) (8–12). However, studies of epigenetic
mechanisms of pain are continuing and reports of epigenetic
modifications on DPNP are currently lacking. Therefore, the present
review aimed to comprehensively summarize the etiology and disease
mechanisms of DPNP.
In the present review, recent studies on epigenetic
modifications in DPNP were summarized. The downstream changes
brought about by epigenetic modifications, including alterations in
the expression levels of ion channels, altered pain-related
receptors, unbalanced inhibitory effects, activation of glial cells
and neuroinflammatory responses were exhaustively presented.
Moreover, the present review assumed that an epigenetics-derived
approach might provide potential diagnosis and therapeutic targets
for DPNP.
Rodent models of DPNP
Rodent models of DM are the source of experimental
evidence for the study and treatment of DPNP and a
mechanism-derived approach might offer a potential
therapeutic-target and an enhanced benefit in attenuating certain
phenotypes of DPNP. The physiology of DPNP in animal models are
similar to humans and the conclusions obtained from different DPNP
animals were introduced in the present review to help understand
the pathogenesis of DPNP and bring some beneficial ideas for the
treatment of DPNP. In addition, as an entire system, animal models
can be used to preliminarily screen a group of potential
therapeutic targets to deal with DPNP and to measure the efficacy
and toxicity of potential drugs. Some in vitro models have
been used to explore specific molecular mechanisms involved in
DPNP, including primary culture of dorsal root ganglion (DRG)
neurons, neural tissue, Schwann cells and other specific cell
lines. Results obtained using in vitro models for the
mechanism of DPNP include, but are not limited to, mitochondrial
events, apoptotic changes and the generation of reactive oxygen
species (5). Although in
vitro models are suitable for investigating molecular
mechanisms of disease, they must ultimately be confirmed in a
living system, that is, an in vivo animal model. Other
aspects of DPNP pathophysiology, especially those involved in pain,
demand the employment of an entire system, necessitating an animal
model. All these rodent models exhibit the three key
characteristics of DPNP in humans: Sensory loss, as measured by
behavioral tests, nerve impairment, measured using
electrophysiology and nerve fiber loss, as detected by the density
of the intraepidermal nerve fiber (IENF). The current models for
DPNP mainly include genetic rodents and streptozotocin (STZ) or
diet-induced approaches.
Genetic models of type 1 diabetes (T1DM)
include the nonobese diabetic and B6Ins2Akita rodents,
while type 2 diabetes (T2DM) models typically use
animals which have a mutation in the leptin gene, known as db/db
rodents. There is currently no sufficient evidence to conclude that
these genetic models are superior to the STZ/diet-induced model in
relation to DPNP (13). In
previously published studies, the STZ/diet-induced model is more
widely used compared with the genetic models. T1DM
occurs due to the destruction of pancreatic β-cells and accounts
for ~5% of DM cases, and is typically associated with autoimmune
issues and a complete insulin deficiency (5,13,14).
The animal model of T1DM is constructed using treatment
with STZ, a cytotoxic methyl nitrosourea substance that can enter
pancreatic β-cells through the glucose 2 transporter, where it
leads to cell death by initiating DNA fragmentation (15). The death of pancreatic β-cells
caused by STZ leads to insulin deficiency and results in the
development of T1DM. Multiple low-dose intraperitoneal
injections of STZ are typically used to initiate a T1DM model in
animals to avoid the toxicity induced by the administration of
excessive levels of STZ. The symptoms of this model initially
present as hyperalgesia in response to noxious stimuli and
eventually develop hypoalgesia and spontaneous pain in the later
stages of disease, which replicates the DPNP symptoms of humans
(16). T2DM can also be
induced by either a high-fat diet alone or a high-fat diet combined
with a single of low-dose STZ (25 mg/kg). Until mice are fed a high
fat diet for 16 weeks, there are no changes in IENF density but
tactile allodynia, thermal hypoalgesia and reduced nerve conduction
velocities are observed (17).
Similar characteristics to patients with prediabetes and obesity
can be observed; however, the spontaneous pain associated with
late-stage DPNP in humans has not been reported in the
high-fat-diet induced animal models of DPNP. Although it may be
challenging to fully replicate the clinical manifestations of human
DPNP in rodent models, these studies are useful tools for studying
the underlying mechanisms of DPNP.
DNA methylation and DPNP
DNA methylation is an epigenetic mechanism involving
the transfer of a methyl group onto the C5 position of cytosine to
form 5-methylcytosine (18). DNA
methylation regulates gene expression by preventing the binding of
transcription factors to DNA or through recruiting certain proteins
involved in gene repression. DNA methylation is performed by DNA
methyltransferases and demethylation by ten-eleven translocation
methylcytosine dioxygenases.
A number of studies have explored the role of DNA
methylation in NP, but there are few reports on the effect of DNA
methylation in DPNP (19–23). In a murine model of DPNP, the
whole-genome bisulfite sequencing of DRGs reveals an altered
methylation pattern in the CpG sites in DNA promoter regions
(24). Additionally, a total of
376 promoter regions with hypermethylated CpG sites and 336
promoter regions with hypomethylated CpG sites were respectively
detected. Furthermore, differentially methylated CpG sites genes
have been reported to be involved in nervous sensory systems and
contribute to the development diabetes or symptoms of pain. A
previous study verified the involvement of one of the
aforementioned genes in DPNP, as it was shown that P2X3R promotes
DNA demethylation and increases interaction with p65, an active
form of NF-κB, contributes to P2X3R sensitization and DPNP
hypersensitivity (25).
Similarity, the expression level of TET2 was increased in a murine
model of DPNP, which resulted in activation of NLRP3 inflammasome
and alleviation of pain sensitivity (26). DNA methylation participates in DPNP
by regulating metabolism and the inflammatory response; however,
there are few studies on the role of DNA methylation in channel
sensitivity, neurodegeneration or dendritic spine remodeling, which
are reported to serve a role in DPNP.
Thus, studies have suggested the potential role of
DNA methylation in DPNP. As there are currently few reports on the
involvement of DNA methylation in DPNP, further studies are
required to elucidate the molecular mechanisms underlying these
epigenetic alterations.
Histone modification and DPNP
The posttranslational modification of histone
proteins is another method of regulation of chromatin structure.
Accumulating evidence indicates that histone modifications
contribute to the occurrence and persistence of NP (27). Histone modification refers to the
process in which histones undergo methylation, acetylation,
phosphorylation, adenylation, ubiquitination, ADP ribosylation and
other modifications by certain enzymes (28,29).
This can alter histone interactions with DNA and other nuclear
proteins, cause the remodeling of chromatin structure and alter
gene expression. Histone acetylation, catalyzed by histone
acetyltransferases, results in the gene transcriptional activation
of genes, whereas histone deacetylation, catalyzed by histone
deacetylases (HDACs), leads to gene repression, which is associated
with the development and maintenance of NP in terms of neuronal
excitability and neuroinflammation in the latest review (30). The review comprehensively shows
that HDACs regulate pivotal genes linked to neuronal activity,
oxidative stress and mitochondrial function in a cellspecific
manner in DRG, which leads to peripheral sensitization in NP and
outlines the recent experimental evidence supporting the
therapeutic potential of HDAC inhibitor in NP. Unlike acetylation,
when acetylation of most histone subunits at any of several lysine
residues can relax chromatin and enhance transcriptional
activation, lysine methylation can cause different effects on gene
expression, depending upon the methylation site (31). For instance, methylation at lysine
4 (K4), K36 and K79 of histone 3 (H3) promotes gene expression,
whereas, methylation at K9 and K27 of H3 is associated with
transcriptional repression (32).
Emerging evidence suggests that enhanced histone
modifications of certain promoters is associated with increased
expression levels of pain-related genes in DPNP. In a rat model of
T1DM, HDAC5 was aberrantly activated in spinal
astrocytes, which promoted STAT3 deacetylation through direct
protein-protein interactions, resulting in a decrease in the
numbers of spinal astrocytes and consequently increasing pain
hypersensitivity (33). Another
histone deacetylase, Sirtuin3 (SIRT3) was also reported to inhibit
oxidative stress and alleviate NP by deacetylating Forkhead box O3a
in the spinal dorsal horn of a diabetic rat model (34). Conversely, reduction in the level
of H3 acetylation in the PDX-1 promoter suppresses the expression
of PDX-1 and inhibits insulin secretion, which may be a potential
cause of both diabetes and DPNP (35). Treatment interventions targeting
histone acetylation may be successful for the treatment of DPNP. A
study reported that HDACs are increased in DPNP and suppression of
HDAC with the HDAC inhibitor FK228 can alleviate diabetic nerve
degeneration and pain (36).
Furthermore, in a murine model of diabetes, valproate sodium serves
a role in decreasing inflammation and pain through inhibition of
spinal HDAC1 expression and glial reactivity (37). However, a clinical trial showed
that the novel HDAC6-inhibitor ricolinostat used for the treatment
of patients with DPNP patients was not associated with a reduction
in NP when compared with placebo (38). Perhaps, despite being called an
HDAC6-inhibitor, unlike other HDAC subtypes, it acts in the cytosol
but not histone and is hypothesized to contribute to the
pathophysiology of small fiber neuropathies in a dose-dependent
manner.
Therefore, the aforementioned studies have
highlighted the role of histone modifications in the
pathophysiological process of DPNP and show that histone
modifications may serve as potential treatment targets for the
management of DPNP. However, despite the various forms of histone
modification, currently, there exists reports only related histone
acetylation modification on DPNP. Other histone modifications in
DPNP such as methylation, phosphorylation, adenylation and
ubiquitination and their potential role in DPNP have yet to be
reported.
RNA modifications and DPNP
The 5′ cap modification and the poly(A) tail of
eukaryotic mRNA are gaining attention for their roles in mRNA
metabolism. The most common internal mRNA modification is
N6-methyladenosine (m6A) and identification of proteins that
recognize, install and remove the marks have highlighted roles for
mRNA modification in numerous aspects of the mRNA life cycle, as
well as in various cellular, developmental and disease processes
(39). The m6A
modification regulate mRNA stability, splicing and translation and
targeting m6A regulatory genes have potential role in
alleviating the pathogenesis of a variety of human diseases.
Methyltransferases, known as m6A ‘writers’, increase
the expression level of m6A. This family of proteins consists of
methyltransferase-like (METTL) 3, METTL14 and Wilms' tumor 1
associated protein (40).
Methyltransferases form a methyltransferase complex and function in
conjunction with each other. These enzymes can be specifically
identified by diverse m6A ‘readers’. The m6A ‘readers’ consist of
YTH domain family 2 and insulin like growth factor 2 mRNA binding
proteins (41). Methylated mRNAs
are recognized by these ‘readers’ and promote m6A methylation
together with methyltransferase. The ‘readers’ may affect NP
through mRNA metabolism via splicing, transcription, subcellular
localization, or degradation. Conversely, m6A-specific
demethylases, AlkB homolog 5 (ALKBH5) and fat-mass and
obesity-associated protein (FTO), can reverse m6A modification of
RNA. The m6A RNA demethylase FTO or ALKBH5, m6A can be converted to
adenosine, which indicates that the process of m6A modification is
dynamically reversible. In addition, other RNA modification such as
N4-Acetylcytidine and RNA-binding proteins (RBPs) serve
diverse roles in the pathophysiology of pain that span RNA
modification in terms of splicing, stability, translation and decay
(42–44).
Previous in vivo and in vitro studies
report that RNA modifications, particularly in m6A, serve crucial
roles in the occurrence and maintenance of NP through regulation of
gene expression, glial cell activation or channel sensitivity in
pain-related areas (20,45–50).
However, the direct research on RNA modification and DPNP is
relatively limited. Similar reports have preliminarily demonstrated
the role of RNA modifications in DPNP, as mRNA splicing alterations
of vascular endothelial growth factor (VEGF) were linked to
neuronal damage associated with DPNP in diabetic rats and humans
and targeting alternative RNA splicing of VEGF could potentially
serve as a new analgesic strategy for the treatment of diabetic
neuropathy (51–54). However, the link between RNA
modifications and mRNA splicing alterations and the mechanisms
regulating mRNA splicing alterations of VEGF are currently unknown
and need further exploration. Similarly, whether RNA modifications
playing a role in DPNP through regulation of gene expression, glial
cell activation or channel sensitivity as in NP is unclear.
Although DPNP, categorized as an NP, is a familiar
complication of diabetes, DPNP has unique characteristics
especially in terms of metabolic abnormalities and the conclusions
obtained from different experimental models of NP may not be fully
applicable to DPNP. Given the significant role of RNA modification
in NP, studies on its potential effects in DPNP is worth further
performed to clarify the molecular events underlying these
epigenetic alterations.
ncRNA and DPNP
ncRNAs are a heterogeneous group of transcripts that
typically do not encode detectable protein, but this does not mean
they are useless or that ncRNAs do not contain information nor have
a function. Compared with mRNAs, the functions of ncRNAs are more
complex and much of the genome can be translated into ncRNAs.
Previous studies have reported that ncRNAs regulate a diverse range
of molecular and biological processes and the aberrant expression
of ncRNAs may be involved with the procession of various human
diseases, including NP (55–63).
A total of three types of ncRNAs have been reported to serve roles
in DPNP: Long non-coding RNA (lncRNA), microRNA (miRNA) and
circular RNA (circRNA). The present review aimed to summarize the
current literature reporting the role of these ncRNAs in DPNP.
Understanding ncRNA expression profiles and their involvement in
certain regulatory networks and the pathogenesis of DPNP may
facilitate the discovery of effective treatments for this
disease.
lncRNA and DPNP
lncRNA are a group of non-coding RNAs >200
nucleotides in length, which are defined as transcripts. lncRNAs
are involved in regulating gene expression at epigenetic,
transcriptional and post-transcriptional levels and their role in
DPNP has been reported (63).
lncRNAs may serve a role as scaffolds for transcriptional and
epigenetic protein complexes, which combine with specific genomic
loci to modulate gene transcription. and also involved in RNA
processing through the direct targeting of mRNAs (64,65).
A microarray analysis with genome-wide expression
patterns of lncRNAs in the spinal dorsal horn identified 1,481
differentially expressed lncRNAs in a murine model of DPNP
(64). Functional analysis showed
that calcium ion transport was the second most significant
biological process of differentially expressed lncRNAs. A total of
289 neighboring and 57 overlapping lncRNA-mRNA pairs, such as
ENSMUST00000150952-Mbp and AK081017-Usp15, may contribute to DPNP
pathogenesis.
In vivo evidence shows certain alleviation of
DPNP with the intervention of specific lncRNAs, which are involved
in epigenetic modifications through affecting purinergic signaling
in the ganglia of diabetic rats (65). In addition, uc.48+ short
interfering (si)RNA treatment in DRG alleviates DPNP by inhibiting
the excitatory transmission mediated by the P2X3 receptor, a member
of an ATP-gated ion channel family (65). Similarly, lncRNA.NONRATT021972
siRNA treatment can repress the upregulated expression and
activation of the P2X3 receptor in DRG and reduce the hyperalgesia
potentiated by the pro-inflammatory cytokine TNF-α in rats with
T2DM (66).
lncRNA-uc.25+ short hairpin (sh)RNA decreases the expression of the
pro-inflammatory P2Y14 receptor, inhibits the release of
inflammatory factors and diminishes the p38 MAPK phosphorylation
(67). DPNP rats treated with
shRNA targeting MSTRG.81401 show an increased mechanical withdrawal
threshold (MWT) and thermal withdrawal latency (TWL), which is
accompanied by decreased expression levels of the P2X4 receptor,
TNF-α and interleukin (IL)-1β in the hippocampus and spinal cord
(68). A previous study reports
that administration of shRNA targeting lncRNA MSTRG.81401 relieves
the hyperalgesia in rats with DPNP (69). NONRATT021972 siRNA treatment
inhibits the expression of P2X7, reduces the release of
inflammatory factors (TNF-α), suppresses the activation of
satellite glial cells (SGCs) in the DRG of rats with
T2DM, restores the excitability of DRG neurons and
ameliorates mechanical and thermal hyperalgesia (70). Another study reports that P2X7
receptor is expressed in SGCs and is involved in hypersensitivity
of DPNP, which is mediated by TRPV1 (71). Rats treated with STZ and siRNA
targeting lncRNA BC168687 show increased MWT and TWL, a decreased
expression level of TRPV1 receptors in DRG and reduced levels of
TNF-α and IL-1β in serum (72).
Intrathecal nerve injury-specific lncRNA antisense oligonucleotides
also mitigate increases in nociceptive hypersensitivity caused by
diabetic neuropathy (73).
In addition, a clinical trial is currently underway
to investigate the role of lncRNA in DPNP. A significantly
increased concentration of lncRNA NONRATT021972 is reported in the
blood and increased severe symptoms of NP are demonstrated in
patients with D2TM (74). lncRNA
NONRATT021972 is positively associated with the severity of DPNP.
In addition, siRNA targeting lncRNA NONRATT021972 attenuates
inflammation through decreasing levels of TNF-α and alleviating
DPNP in a murine model.
In summary, there are increasing reports that
suggest the involvement of lncRNAs in DPNP. However, current
studies on lncRNA and its association with DPNP lack depth. Current
reports mainly focus on the mechanism of action of the
lncRNA/miRNA/target axis or treatments targeting lncRNAs. However,
lncRNAs may directly bind to ligands, receptors or enzymes and be
involved in the pathogenesis of DPNP (75).
miRNA and DPNP
miRNAs are endogenous, single-stranded RNAs ranging
from 21–25 nucleotides in length. miRNAs regulate gene expression
and >60% of genes are targeted by miRNAs (76). The aberrant activity of miRNAs
contributes to various complications in diabetes, including
nephropathy, retinopathy and DPNP. miRNAs regulate various
processes involved in diabetic complications related to
inflammation, provoking pain and metabolic syndrome pathways
(77). Furthermore, a number of
miRNAs are implicated in the process of DPNP (78). Therefore, various miRNAs and their
targets may serve as potential biomarkers for the diagnosis,
prognosis and therapeutic interventions of DPNP.
RNA sequencing of a DPNP rodents model show
alterations in miRNA expression levels and their regulatory
networks. An miRNA array profile of DRGs of mice with
T1DM shows that miRNA (miR)-33 and miR-380 expressed in
nociceptive neurons are critical determinants of diabetic pain and
miR-124-1 is an important mediator for physiological nociception
(79). Further functional analyses
identified variant functional roles for four miRNAs that are
prominently dysregulated in DPNP, namely that overexpression of i)
pre-miR-33 alleviates mechanical hypersensitivity, ii) pre-miR-380
enhances mechanical hypersensitivity, iii) pre-miR-124 increases
basal mechanical sensitivity without involving in diabetes-induced
mechanical hypersensitivity and iv) pre-miRNA-335 serves no role in
either basal or diabetes-induced mechanical hypersensitivity.
However, another study reports that miR-122 is the most notably
upregulated miRNA as determined by sequencing analysis of
STZ-induced mice (80) There is an
association between miR-122 and NP (81); however, whether miR-122 is involved
in DPNP remains unclear and requires further exploration.
The expression levels of certain miRNAs in different
models of DPNP and their targets are inconsistent. The expression
levels of some miRNAs are negatively correlated with the occurrence
and persistence of DPNP. miR-193a expression is decreased in the
lumbar spinal dorsal horn of STZ-induced diabetic mice and
overexpression of miR-193a alleviates DPNP through the inhibition
of HMGB1 expression (82).
miR-184-5p expression is decreased in spinal dorsal in diabetic
mice, while intrathecal injection of miR-184-5p agomir attenuates
DPNP; by contrast, intrathecal miR-184-5p antagomir exacerbates
pain-associated behaviors (83).
Low concentrations of bupivacaine inhibits microglial inflammation
and attenuates diabetic neuropathy through downregulating PDE4B via
activating miR-23a expression, indicating the presence of a
negative relationship between pain and miRNA-23a expression
(84). miR-96 expression, reversed
by participating in swimming exercises, alleviates pain associated
with diabetes by inhibiting the expression of NaV1.3 in
rats with T2DM, indicating the analgesic potential of
miR-96 in DPNP (85). miR-145
expression precludes the development of mechanical hyperalgesia via
suppression of Nav1.8 in diabetic rats (86). Furthermore, the expression level of
miR-590-3p is decreased in mice with DPNP and miR-590-3p agomir
ameliorates pain-related behavior, reduces expression of the
inflammatory cytokines TNF-α, IL-1β and IL-6 and suppresses neural
infiltration by immune cells in db/db mice (87). Similarly, miR-497 alleviates DPNP,
which is associated with suppression of USP15, a reported target of
miR-497 (88). miR-503-5p
alleviates peripheral neuropathy-induced pain in T2DM
mice by regulating SEPT9 expression (89). Glucose-induced astrocytes exhibit a
decrease in miR-125a-5p expression levels and administration of the
miR-125a-5p mimic in db/db mice attenuated DPNP (90). In a number of clinical trials, the
expression levels of miR-1-3p and miR-199a-3p were decreased and
increased, respectively, in whole blood samples from patients with
diabetic neuropathy compared with the healthy controls, suggesting
their diverse and potential relationship with progression of DPNP
(91,92).
By contrast, the expression of some miRNAs are
positively associated with the occurrence and persistence of DPNP.
Inhibition of miR-221 can reduce pain and decrease expression of
inflammatory factors by targeting SOCS3 in DPNP (93). In a rat model of DM, miR-133a-3p
antagomir administration alleviates DPNP and downregulates p-p38
phosphorylation and overexpression of miR-133a-3p in sciatic nerve
induced pain (94). miR-155
targets and suppresses Nrf2 expression in DPNP and miR-155
silencing improves angiogenesis and alleviates inflammation and
sciatic nerve injury in DPNP rats (95). Subcutaneous injection of the
miRNA-expressing HSV vector targeting NaVα subunits into
the feet of a diabetic rats models to transduce DRG leads to a
decrease in the expression of NaVα subunits l in DRG
neurons, which was associated with an alleviation of symptoms such
as cold allodynia, thermal hyperalgesia and mechanical hyperalgesia
(96).
Therefore, manipulation of miRNA expression levels
is an emerging diagnostic and therapeutic approach in which to
screen and identify nerve-specific targets or successful drug
delivery systems. This will be vital to ensure suitable safety
profiles that could facilitate the translation of this technology
to future clinical use.
circRNA and DPNP
Inconsistent with linear RNAs, circular (circ)RNAs
are circular molecules with covalently closed loop structures that
are exempted from exonucleases. These stable characteristics of
circRNAs enable them to be highly expressed in both cells and
extracellular vesicles and they are expressed at different levels
during the process of diseases (97). As a type of ncRNA, circRNAs exert
biological functions by acting as transcriptional regulators,
protein templates and miRNA sponges that thereby affect the
expression levels of miRNAs and their target genes, circRNAs also
serve as decoys or sponges of RNA-binding proteins that influence
the expression and function of coding mRNAs and are associated with
various biological processes including NP (98,99).
There are currently few reports which evaluate the
role and mechanisms of circRNAs in DPNP. An analysis of
high-throughput RNA sequencing data of DRGs comparing wild-type
mice and mice with diabetic neuropathy shows that 11 circRNAs and
14 mRNAs have a significant correlation and the expression of
circRNA.4614 is clearly upregulated, suggesting that such
dysregulated circular RNAs may be involved in the initiation and
progression of DPNP (98). Another
sequencing analysis identifies 135 different expression circRNAs
(64 upregulated and 71 downregulated) in spinal cord between the
DPNP and control group, but with no further functional verification
(80). circHIPK3 is highly
expressed in serum samples obtained from diabetes patients with
DPNP and in DRG from STZ-induced diabetic rats (100). Additionally, the expression level
of circHIPK3 is positively associated with the grade of NP in
patients with T2DM. In diabetic rats, downregulating
circHIPK3 attenuates NP. Further study demonstrates that circHIPK3
target miRNA-124 and suppress its expression level to reverse the
inhibitory effects of miRNA-124 on neuroinflammation. The
aforementioned in vivo and clinical study suggests that
circRNA HIPK3 may act as a potential target for the treatment of
DPNP. However, further studies are required to explore the
therapeutic potential for targeting dysregulated circRNAs to treat
DPNP.
In conclusion, ncRNAs may serve as a promising
biomolecule for treatment of DPNP as epigenetic modifications of
ncRNAs contributes to the pathophysiology of this condition
(Table I). A number of
dysregulated ncRNAs have been reported in the serum of patients
with DPNP, which can be considered as potential biomarkers for
early diagnosis of this condition. As certain ncRNAs have shown
promise for alleviating DPNP in in vivo experiments,
treatment targeting these ncRNAs may be a promising therapeutic
approach for patients with DPNP in the future.
 | Table I.Non-coding RNA related to diabetic
peripheral neuropathic pain. |
Table I.
Non-coding RNA related to diabetic
peripheral neuropathic pain.
| Epigenetic
modifications | Epigenetic
effect | Region | Target | Result | (Refs.) |
|---|
| lncRNA | lncRNA uc.48+ | DRG | P2X3, ERK1/2 | Mechanical
allodynia and thermal hyperalgesia | (65) |
|
| lncRNA | DRG | P2X3, ERK1/2 | Mechanical
allodynia and thermal hyperalgesia | (66) |
|
| NONRATT021972
↑ |
|
|
|
|
|
| lncRNA-UC.25 +
↑ | Spinal | STAT1, | Activation of
microglia, increasing the | (67) |
|
|
| cord | P2Y14 | expression of
inflammatory factors and the |
|
|
|
|
| receptor | level of p38
mitogen-activated protein kinase |
|
|
|
|
|
|
phosphorylation |
|
|
| lncRNA | Hippocampus, | P2X4 receptor, | TNF-α, and IL-1β,
mechanical allodynia and | (68, |
|
| MSTRG.81401 ↑ | spinal cord | ERK1/2, P2X7 | thermal
hyperalgesia, depression-like behaviors | 69) |
|
|
|
| receptor |
|
|
|
| lncRNA | DRG | P2X7 receptor | TNF-α expression,
mechanical allodynia and | (70) |
|
| NONRATT021972
↑ |
|
| thermal
hyperalgesia |
|
|
| lncRNA BC168687
↑ | DRG | TRPV1 | Mechanical
allodynia and thermal hyperalgesia | (72) |
|
| NIS-lncRNA/ | DRG | CCL2 | Nociceptive
hypersensitivity | (73) |
|
| lncRNA | Blood | TNF-α | Mechanical
allodynia and thermal hyperalgesia | (74) |
|
| NONRATT021972
↑ |
|
|
|
|
| miRNA | miR-33 ↑ miR-380
↑ | DRG | A set of | Mechanical
hypersensitivity | (79) |
|
| miR-124 ↑ |
| mRNAs |
|
|
|
| miR-122 ↑ | Spinal cord | A set of | Neuropathic
pain | (80) |
|
|
|
| mRNAs |
|
|
|
| miR-193a ↓ | Spinal dorsal
horn | HMGB1 | Peripheral
neuroinflammation, neuropathic pain | (82) |
|
| miR-184-5p ↓ | Spinal dorsal
horn | CCL1 | Mechanical
hypersensitivity | (83) |
|
| miR-23a ↓ | Spinal cord | PDE4B | inflammatory
cytokines, mechanical allodynia and thermal hyperalgesia | (84) |
|
| miR-96 ↓ | Sciatic nerve |
NaV1.3 | Thermal
hyperalgesia | (85) |
|
| miR-145 ↓ | DRG | Nav1.8 | Mechanical
hyperalgesia | (86) |
|
| miR-590-3p ↓ | DRG | RAP1A | T cells
proliferation and migration, mechanical allodynia and thermal
hyperalgesia | (87) |
|
| miR-497 ↓ | DRG | USP15 | G6PD expression,
mechanical allodynia and thermal hyperalgesia | (88) |
|
| miR-503-5p ↓ | Spinal cord | SEPT9 | Astrocyte
activation, mechanical allodynia and thermal hyperalgesia | (89) |
|
| miR-125a-5p ↓ | Sciatic nerve | TRAF6 | Astrocyte
activation, mechanical allodynia and thermal hyperalgesia | (90) |
|
| miR-1-3p ↓ | Blood mononuclear
cells | CXCR4 | Patients' diabetic
neuropathy | (91) |
|
| miR-199a-3p ↑ | Lower limb skin
tissues, plasma of peripheral blood | SerpinE2 | Patients' diabetic
neuropathy | (92) |
|
| miR-221 ↑ | Serum exosomes | SOCS3 | Expression of
inflammatory factors, mechanical allodynia and thermal
hyperalgesia | (93) |
|
| miR-133a-3p ↑ | Sciatic nerve | p38 | Mechanical
allodynia, p-p38 MAPK activation | (94) |
|
| miR-155 ↑ | Sciatic nerve | Nrf2 | Sciatic nerve
injury, inflammation | (95) |
| cicRNA | circRNA.4614 ↑ | DRG | A set of | Sciatic nerve
injury, mechanical allodynia and | (98) |
|
| other 11 |
| mRNAs | thermal
hyperalgesia |
|
|
| circRNAs ↑ |
|
|
|
|
|
| 64 circRNAs ↑ | Spinal cord | / | Mechanical
allodynia and thermal hyperalgesia | (80) |
|
| 71 circRNAs ↓ |
|
|
|
|
|
| circHIPK3 ↑ | Serum | miR-124 | Neuroinflammation,
mechanical allodynia and | (100) |
|
|
| (patients) |
| thermal
hyperalgesia |
|
|
|
| DRG (rats) |
|
|
|
Conclusions and future perspectives
The present review identified the role of epigenetic
modifications in DPNP. The roles of different forms of epigenetic
modifications, including DNA methylation, histone modification, RNA
modification and ncRNA, were summarized to demonstrate their
potential effects on regulating the occurrence and maintenance of
DPNP (Fig. 1). The experimental
approach of targeting these epigenetic modifications shows
potential for the alleviation of DPNP in diabetic models. Despite
current evidence highlighting the role of epigenetic modifications
in NP, there are limited reports on a number of forms of the
epigenetic modifications involved in DPNP, particularly RNA
modifications, which need to be further studied and requires more
evidence in the future to verify the role of RNA modification in
DPNP. In addition, there are too few studies on the effect of
epigenetic modifications on the neural circuit to draw solid
conclusions. Activating certain neural circuits can alleviate pain
and its associated comorbidities, such as depression and anxiety,
while activating other circuits can exacerbate these conditions
(101–103). An increasing number of studies
research the role of neural circuits in pain; however, the
relationship between neural circuits and epigenetic modifications
in DPNP is currently ambiguous and requires further exploration.
Additionally, the mechanisms of action of epigenetic modifications
in DPNP have mainly been derived from rodent studies and few of the
conclusions drawn from such studies have been verified and applied
in a clinical setting. Finally, the upstream mechanisms of
epigenetic modifications are unclear at present. For example,
whether the changes in the internal homeostatic environment in DM
will promote the epigenetic alterations through post-translational
modifications, namely O-GlcNAcylation, to regulate the development
of DPNP, remains to be further studied. Further research will be
required to develop the results of mechanistic studies into
clinically available treatments.
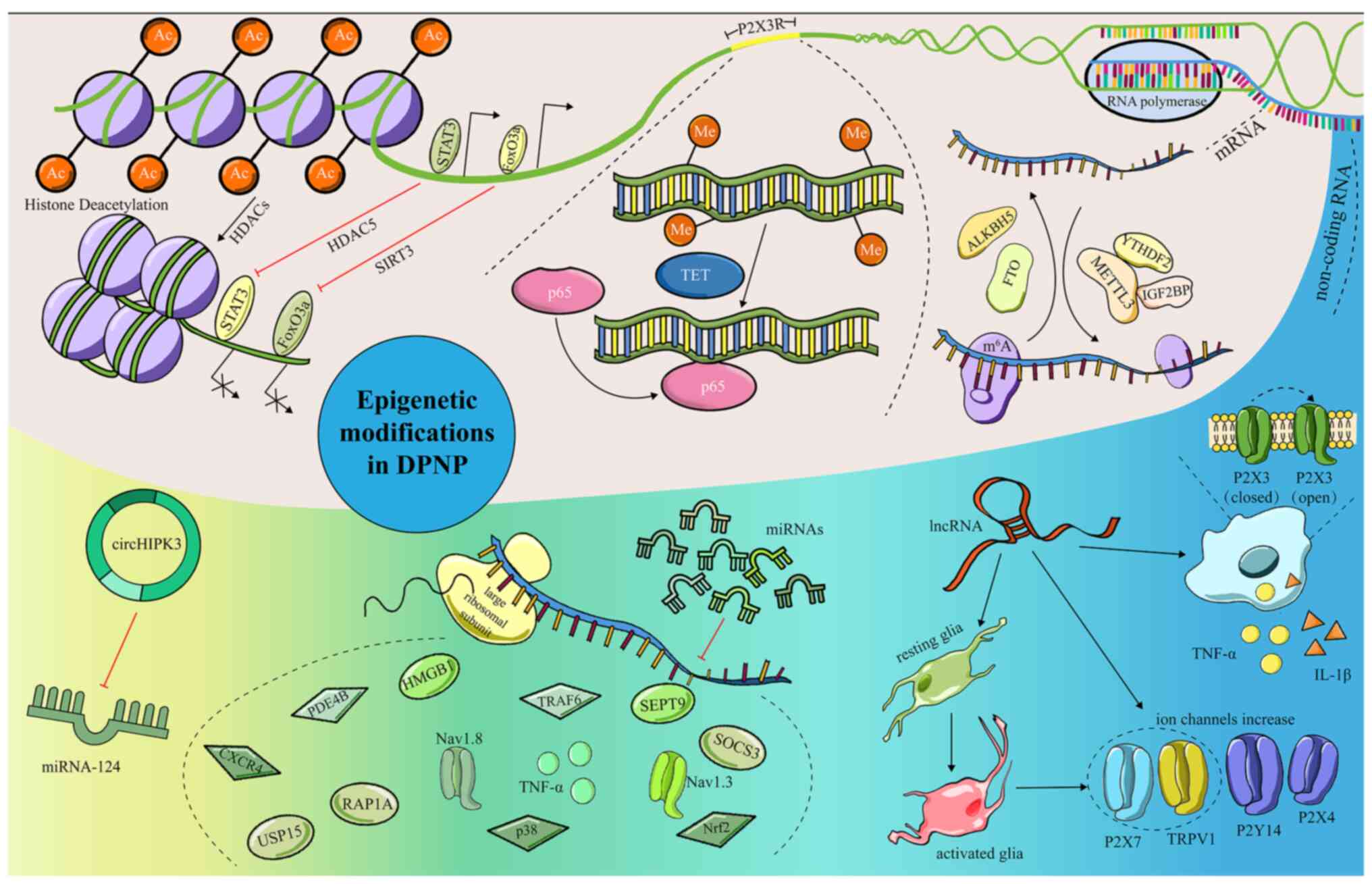 | Figure 1.Epigenetic modifications and their
regulatory mechanism in DPNP. Epigenetic modifications can
participate in pain regulation through various mechanisms. Some
common and key mechanisms mainly include pain-related signaling
pathways; pain-related receptors and targets; regulation of
cytokines release; and targeting ion channels related to pain.
Epigenetic modifications that are involved in the regulation of
DPNP include (A) Histone modification, causing the remodeling of
chromatin structure and alter gene expression, (B) DNA methylation,
regulating gene expression, (C) RNA modification, involving in mRNA
metabolism and (D) ncRNA, serving diverse roles in pain. DPNP,
diabetic peripheral neuropathic pain; HDAC, histone deacetylase;
TETs, methylcytosine dioxygenases; ALKBH5, AlkB homolog 5; FTO,
fat-mass and obesity-associated protein; METTL3,
methyltransferase-like 3; YTHDF2, YTH domain family 2; IGF2BP,
insulin like growth factor 2 mRNA binding proteins; miRNA,
microRNA; circ, circularRNA; FoxO3a, Forkhead box O3a; SIRT3,
Sirtuin3; IL, interleukin. |
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of
Sichuan (grant no. 2022NSFSC0672) and hospital level project of the
General Hospital of the Western Theater Command (Sichuan, China;
grant no. 2021-XZYG-B15).
Availability of data and materials
Not applicable.
Authors' contributions
TG and JL wrote the manuscript. HY designed and
wrote the manuscript. JF and GG revised the manuscript and drew the
table and figure. Data authentication is not applicable. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Behl T, Gupta A, Sehgal A, Sharma S, Singh
S, Sharma N, Diaconu CC, Rahdar A, Hafeez A, Bhatia S, et al: A
spotlight on underlying the mechanism of AMPK in diabetes
complications. Inflamm Res. 70:939–957. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abbott CA, Malik RA, van Ross ERE,
Kulkarni J and Boulton AJM: Prevalence and characteristics of
painful diabetic neuropathy in a large community-based diabetic
population in the U.K. Diabetes Care. 34:2220–2224. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feldman EL, Nave KA, Jensen TS and Bennett
DLH: New horizons in diabetic neuropathy: Mechanisms,
bioenergetics, and pain. Neuron. 93:1296–1313. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sloan G, Selvarajah D and Tesfaye S:
Pathogenesis, diagnosis and clinical management of diabetic
sensorimotor peripheral neuropathy. Nat Rev Endocrinol. 17:400–420.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frank T, Nawroth P and Kuner R:
Structure-function relationships in peripheral nerve contributions
to diabetic peripheral neuropathy. Pain. 160 (Suppl 1):S29–S36.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tesfaye S, Sloan G, Petrie J, White D,
Bradburn M, Julious S, Rajbhandari S, Sharma S, Rayman G, Gouni R,
et al: Comparison of amitriptyline supplemented with pregabalin,
pregabalin supplemented with amitriptyline, and duloxetine
supplemented with pregabalin for the treatment of diabetic
peripheral neuropathic pain (OPTION-DM): A multicentre,
double-blind, randomised crossover trial. Lancet. 400:680–690.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berger SL, Kouzarides T, Shiekhattar R and
Shilatifard A: An operational definition of epigenetics. Genes Dev.
23:781–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang X, Shen X, Xu Y, Xu S, Xia F, Zhu B,
Liu Y, Wang W, Wu H and Wang F: The etiological changes of
acetylation in peripheral nerve injury-induced neuropathic
hypersensitivity. Mol Pain. 14:17448069187984082018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khangura RK, Bali A, Jaggi AS and Singh N:
Histone acetylation and histone deacetylation in neuropathic pain:
An unresolved puzzle? Eur J Pharmacol. 795:36–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lutz BM, Bekker A and Tao YX: Noncoding
RNAs: New players in chronic pain. Anesthesiology. 121:409–417.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang L, Lutz BM, Bekker A and Tao YX:
Epigenetic regulation of chronic pain. Epigenomics. 7:235–245.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo D, Li X, Tang S, Song F, Li W, Xie G,
Liang J and Zhou J: Epigenetic modifications in neuropathic pain.
Mol Pain. 17:174480692110567672021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
O'Brien PD, Sakowski SA and Feldman EL:
Mouse models of diabetic neuropathy. ILAR J. 54:259–272. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Devendra D, Liu E and Eisenbarth GS: Type
1 diabetes: Recent developments. BMJ. 328:750–754. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eleazu CO, Eleazu KC, Chukwuma S and
Essien UN: Review of the mechanism of cell death resulting from
streptozotocin challenge in experimental animals, its practical use
and potential risk to humans. J Diabetes Metab Disord. 12:602013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agarwal N, Helmstädter J, Rojas DR, Bali
KK, Gangadharan V and Kuner R: Evoked hypoalgesia is accompanied by
tonic pain and immune cell infiltration in the dorsal root ganglia
at late stages of diabetic neuropathy in mice. Mol Pain.
14:17448069188179752018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Obrosova IG, Ilnytska O, Lyzogubov VV,
Pavlov IA, Mashtalir N, Nadler JL and Drel VR: High-fat diet
induced neuropathy of pre-diabetes and obesity: Effects of
‘healthy’ diet and aldose reductase inhibition. Diabetes.
56:2598–2608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun L, Gu X, Pan Z, Guo X, Liu J, Atianjoh
FE, Wu S, Mo K, Xu B, Liang L, et al: Contribution of DNMT1 to
neuropathic pain genesis partially through epigenetically
repressing Kcna2 in primary afferent neurons. J Neurosci.
39:6595–6607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Zhao X, Wang J, Jin Y, Gong M, Ye
Y and Li P: METTL3 suppresses neuropathic pain via modulating
N6-methyladenosine-dependent primary miR-150 processing. Cell Death
Discov. 8:802022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jang JH, Song EM, Do YH, Ahn S, Oh JY,
Hwang TY, Ryu Y, Jeon S, Song MY and Park HJ: Acupuncture
alleviates chronic pain and comorbid conditions in a mouse model of
neuropathic pain: The involvement of DNA methylation in the
prefrontal cortex. Pain. 162:514–530. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Z, Tao W, Hou YY, Wang W, Kenny PJ
and Pan ZZ: MeCP2 repression of G9a in regulation of pain and
morphine reward. J Neurosci. 34:9076–9087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weng YL, An R, Cassin J, Joseph J, Mi R,
Wang C, Zhong C, Jin SG, Pfeifer GP, Bellacosa A, et al: An
intrinsic epigenetic barrier for functional axon regeneration.
Neuron. 94:337–346.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen W, Lan T, Sun Q, Zhang Y, Shen D, Hu
T, Liu J, Chong Y, Wang P, Li Q, et al: Whole genomic DNA
methylation profiling of CpG sites in promoter regions of dorsal
root ganglion in diabetic neuropathic pain mice. J Mol Neurosci.
71:2558–2565. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang HH, Hu J, Zhou YL, Qin X, Song ZY,
Yang PP, Hu S, Jiang X and Xu GY: Promoted interaction of nuclear
factor-κB with demethylated purinergic P2X3 receptor gene
contributes to neuropathic pain in rats with diabetes. Diabetes.
64:4272–4284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen W, Wang X, Sun Q, Zhang Y, Liu J, Hu
T, Wu W, Wei C, Liu M, Ding Y, et al: The upregulation of NLRP3
inflammasome in dorsal root ganglion by ten-eleven translocation
methylcytosine dioxygenase 2 (TET2) contributed to diabetic
neuropathic pain in mice. J Neuroinflammation. 19:3022022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Penas C and Navarro X: Epigenetic
modifications associated to neuroinflammation and neuropathic pain
after neural trauma. Front Cell Neurosci. 12:1582018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bannister AJ and Kouzarides T: Regulation
of chromatin by histone modifications. Cell Res. 21:381–395. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang W, Jiao B, Yu S, Zhang C, Zhang K,
Liu B and Zhang X: Histone deacetylase as emerging pharmacological
therapeutic target for neuropathic pain: From epigenetic to
selective drugs. CNS Neurosci Ther. 30:e147452024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Morales S, Monzo M and Navarro A:
Epigenetic regulation mechanisms of microRNA expression. Biomol
Concepts. 8:203–212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qi R, Cao J, Sun Y, Li Y, Huang Z, Jiang
D, Jiang XH, Snutch TP, Zhang Y and Tao J: Histone
methylation-mediated microRNA-32-5p down-regulation in sensory
neurons regulates pain behaviors via targeting Cav3.2 channels.
Proc Natl Acad Sci USA. 119:e21172091192022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fan T, Yu Y, Chen YL, Gu P, Wong S, Xia
ZY, Liu JA and Cheung CW: Histone deacetylase 5-induced deficiency
of signal transducer and activator of transcription-3 acetylation
contributes to spinal astrocytes degeneration in painful diabetic
neuropathy. Glia. 71:1099–1119. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou C, Zhang Y, Jiao X, Wang G, Wang R
and Wu Y: SIRT3 alleviates neuropathic pain by deacetylating FoxO3a
in the spinal dorsal horn of diabetic model rats. Reg Anesth Pain
Med. 46:49–56. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding H, Wang F, Shi X, Ma H, Du Y, Hou L
and Xing N: LncRNA MALAT1 induces the dysfunction of β cells via
reducing the histone acetylation of the PDX-1 promoter in type 1
diabetes. Exp Mol Pathol. 114:1044322020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thakur V, Gonzalez MA, Parada M, Martinez
RD and Chattopadhyay M: Role of histone deacetylase inhibitor in
diabetic painful neuropathy. Mol Neurobiol. 61:2283–2296. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Elsherbiny NM, Ahmed E, Kader GA,
Abdel-Mottaleb Y, ElSayed MH, Youssef AM and Zaitone SA: Inhibitory
effect of valproate sodium on pain behavior in diabetic mice
involves suppression of spinal histone deacetylase 1 and
inflammatory mediators. Int Immunopharmacol. 70:16–27. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Michelson D, Chin WW, Dworkin RH, Freeman
R, Herrmann DN, Mazitschek R, Pop-Busui R, Shaibani A, Vornov J,
Jones M, et al: A randomized, double-blind, placebo-controlled
study of histone deacetylase type 6 inhibition for the treatment of
painful diabetic peripheral neuropa. Pain Rep. 8:e11142023.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bokar JA, Shambaugh ME, Polayes D, Matera
AG and Rottman FM: Purification and cDNA cloning of the
AdoMet-binding subunit of the human mRNA
(N6-adenosine)-methyltransferase. RNA. 3:1233–1247. 1997.PubMed/NCBI
|
|
41
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Smith PR and Campbell ZT: RNA-binding
proteins in pain. Wiley Interdiscip Rev RNA. 15:e18432024.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu T, Wang J, Wu Y, Wu JY, Lu WC, Liu M,
Zhang SB, Xie D, Xin WJ and Xie JD: Ac4C enhances the translation
efficiency of vegfa mRNA and mediates central sensitization in
spinal dorsal horn in neuropathic pain. Adv Sci (Weinh).
10:e23031132023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Q, Weng W, Gu X, Xiang J, Yang Y,
Zhu MX, Gu W, He Z and Li Y: hnRNPA1 SUMOylation promotes cold
hypersensitivity in chronic inflammatory pain by stabilizing TRPA1
mRNA. Cell Rep. 42:1134012023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang Z, Zhang Y, Wang S, Qi R, Tao Y, Sun
Y, Jiang D, Jiang X and Tao J: FOXD3-mediated transactivation of
ALKBH5 promotes neuropathic pain via m6A-dependent
stabilization of 5-HT3A mRNA in sensory neurons. Proc Natl Acad Sci
USA. 121:e23128611212024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lu W, Yang X, Zhong W, Chen G, Guo X, Ye
Q, Xu Y, Qi Z, Ye Y, Zhang J, et al: METTL14-mediated m6A
epitranscriptomic modification contributes to chemotherapy-induced
neuropathic pain by stabilizing GluN2A expression via IGF2BP2. J
Clin Invest. 134:e1748472024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang K, Li P, Jia Y, Liu M and Jiang J:
Non-coding RNA and n6-methyladenosine modification play crucial
roles in neuropathic pain. Front Mol Neurosci. 15:10020182022.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang XL, Wei X, Yuan JJ, Mao YY, Wang ZY,
Xing N, Gu HW, Lin CH, Wang WT, Zhang W and Xing F: Downregulation
of fat mass and obesity-related protein in the anterior cingulate
cortex participates in anxiety- and depression-like behaviors
induced by neuropathic pain. Front Cell Neurosci. 16:8842962022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zeng F, Cao J, Hong Z, Lu Y, Qin Z and Tao
T: Epigenetic combined with transcriptomic analysis of the m6A
methylome after spared nerve injury-induced neuropathic pain in
mice. Neural Regen Res. 18:2545–2552. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang W, Qiao SC, Wu XB, Sun B, Yang JG, Li
X, Zhang X, Qian SJ, Gu YX and Lai HC: Circ_0008542 in osteoblast
exosomes promotes osteoclast-induced bone resorption through m6A
methylation. Cell Death Dis. 12:6282021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hulse RP, Beazley-Long N, Ved N, Bestall
SM, Riaz H, Singhal P, Ballmer Hofer K, Harper SJ, Bates DO and
Donaldson LF: Vascular endothelial growth factor-A165b prevents
diabetic neuropathic pain and sensory neuronal degeneration. Clin
Sci (Lond). 129:741–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Quattrini C, Jeziorska M, Boulton AJM and
Malik RA: Reduced vascular endothelial growth factor expression and
intra-epidermal nerve fiber loss in human diabetic neuropathy.
Diabetes Care. 31:140–145. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hulse RP, Beazley-Long N, Hua J, Kennedy
H, Prager J, Bevan H, Qiu Y, Fernandes ES, Gammons MV,
Ballmer-Hofer K, et al: Regulation of alternative VEGF-A mRNA
splicing is a therapeutic target for analgesia. Neurobiol Dis.
71:245–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bestall SM, Hulse RP, Blackley Z, Swift M,
Ved N, Paton K, Beazley-Long N, Bates DO and Donaldson LF: Sensory
neuronal sensitisation occurs through HMGB-1-RAGE and TRPV1 in
high-glucose conditions. J Cell Sci. 131:jcs2159392018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Diederichs S: Non-coding RNA and disease.
RNA Biol. 9:701–702. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Good DJ: Non-coding RNAs in human health
and diseases. Genes (Basel). 14:14292023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li C, Ni YQ, Xu H, Xiang QY, Zhao Y, Zhan
JK, He JY, Li S and Liu YS: Roles and mechanisms of exosomal
non-coding RNAs in human health and diseases. Signal Transduct
Target Ther. 6:3832021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nemeth K, Bayraktar R, Ferracin M and
Calin GA: Non-coding RNAs in disease: From mechanisms to
therapeutics. Nat Rev Genet. 25:211–232. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roganović J and Petrović N: Clinical
perspectives of non-coding RNA in oral inflammatory diseases and
neuropathic pain: A narrative review. Int J Mol Sci. 23:82782022.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jiang BC, Cao DL, Zhang X, Zhang ZJ, He
LN, Li CH, Zhang WW, Wu XB, Berta T, Ji RR and Gao YJ: CXCL13
drives spinal astrocyte activation and neuropathic pain via CXCR5.
J Clin Invest. 126:745–761. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang B, Ma L, Guo X, Du S, Feng X, Liang
Y, Govindarajalu G, Wu S, Liu T, Li H, et al: A sensory
neuron-specific long non-coding RNA reduces neuropathic pain by
rescuing KCNN1 expression. Brain. 146:3866–3884. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu M, Feng Y and Shi X: Advances with long
non-coding RNAs in diabetic peripheral neuropathy. Diabetes Metab
Syndr Obes. 13:1429–1434. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Du H, Liu Z, Tan X, Ma Y and Gong Q:
Identification of the genome-wide expression patterns of long
non-coding RNAs and mRNAs in mice with streptozotocin-induced
diabetic neuropathic pain. Neuroscience. 402:90–103. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang S, Xu H, Zou L, Xie J, Wu H, Wu B, Yi
Z, Lv Q, Zhang X, Ying M, et al: LncRNA uc.48+ is involved in
diabetic neuropathic pain mediated by the P2X3 receptor in the
dorsal root ganglia. Purinergic Signal. 12:139–148. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Peng H, Zou L, Xie J, Wu H, Wu B, Zhu G,
Lv Q, Zhang X, Liu S, Li G, et al: lncRNA NONRATT021972 siRNA
decreases diabetic neuropathic pain mediated by the P2X3
receptor in dorsal root ganglia. Mol Neurobiol. 54:511–523. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wu B, Zhou C, Xiao Z, Tang G, Guo H, Hu Z,
Hu Q, Peng H, Pi L, Zhang Z, et al: LncRNA-UC.25 + shRNA alleviates
P2Y14 receptor-mediated diabetic neuropathic pain via
STAT1. Mol Neurobiol. 59:5504–5515. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sun M, Zhang M, Yin H, Tu H, Wen Y, Wei X,
Shen W, Huang R, Xiong W, Li G and Gao Y: Long non-coding RNA
MSTRG.81401 short hairpin RNA relieves diabetic neuropathic pain
and behaviors of depression by inhibiting P2X4 receptor expression
in type 2 diabetic rats. Purinergic Signal. 19:123–133. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhan T, Tang S, Du J, Liu J, Yu B, Yang Y,
Xie Y, Qiu Y, Li G and Gao Y: Implication of lncRNA MSTRG.81401 in
hippocampal pyroptosis induced by P2X7 receptor in type 2 diabetic
rats with neuropathic pain combined with depression. Int J Mol Sci.
25:11862024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu S, Zou L, Xie J, Xie W, Wen S, Xie Q,
Gao Y, Li G, Zhang C, Xu C, et al: LncRNA NONRATT021972 siRNA
regulates neuropathic pain behaviors in type 2 diabetic rats
through the P2X7 receptor in dorsal root ganglia. Mol Brain.
9:442016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang A, Shi X, Yu R, Qiao B, Yang R and Xu
C: The P2X7 receptor is involved in diabetic neuropathic
pain hypersensitivity mediated by TRPV1 in the rat dorsal root
ganglion. Front Mol Neurosci. 14:6636492021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu C, Li C, Deng Z, Du E and Xu C: Long
non-coding RNA BC168687 is involved in TRPV1-mediated diabetic
neuropathic pain in rats. Neuroscience. 374:214–222. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wen CH, Berkman T, Li X, Du S,
Govindarajalu G, Zhang H, Bekker A, Davidson S and Tao YX: Effect
of intrathecal NIS-lncRNA antisense oligonucleotides on neuropathic
pain caused by nerve trauma, chemotherapy, or diabetes mellitus. Br
J Anaesth. 130:202–216. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yu W, Zhao GQ, Cao RJ, Zhu ZH and Li K:
LncRNA NONRATT021972 was associated with neuropathic pain scoring
in patients with type 2 diabetes. Behav Neurol. 2017:29412972017.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang C, Gao R, Zhou R, Chen H, Liu C, Zhu
T and Chen C: The emerging power and promise of non-coding RNAs in
chronic pain. Front Mol Neurosci. 15:10379292022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kaur P, Kotru S, Singh S and Munshi A:
Role of miRNAs in diabetic neuropathy: Mechanisms and possible
interventions. Mol Neurobiol. 59:1836–1849. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Meydan C, Üçeyler N and Soreq H:
Non-coding RNA regulators of diabetic polyneuropathy. Neurosci
Lett. 731:1350582020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bali KK, Gandla J, Rangel DR, Castaldi L,
Mouritzen P, Agarwal N, Schmelz M, Heppenstall P and Kuner R: A
genome-wide screen reveals microRNAs in peripheral sensory neurons
driving painful diabetic neuropathy. Pain. 162:1334–1351. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
He J, Wang HB, Huang JJ, Zhang L, Li DL,
He WY, Xiong QM and Qin ZS: Diabetic neuropathic pain induced by
streptozotocin alters the expression profile of non-coding RNAs in
the spinal cord of mice as determined by sequencing analysis. Exp
Ther Med. 22:7752021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ma F, Wang C, Yoder WE, Westlund KN,
Carlson CR, Miller CS and Danaher RJ: Efficacy of herpes simplex
virus vector encoding the human preproenkephalin gene for treatment
of facial pain in mice. J Oral Facial Pain Headache. 30:42–50.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu B, Guo Y, Chen Q, Xiong Q and Min S:
MicroRNA-193a downregulates HMGB1 to alleviate diabetic neuropathic
pain in a mouse model. Neuroimmunomodulation. 26:250–257. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wu D, Zhong S, Du H, Han S, Wei X and Gong
Q: MiR-184-5p represses neuropathic pain by regulating CCL1/CCR8
signaling interplay in the spinal cord in diabetic mice. Neurol
Res. 46:54–64. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang X, Xia L, Xie A, Liao O, Ju F and
Zhou Y: Low concentration of bupivacaine ameliorates painful
diabetic neuropathy by mediating miR-23a/PDE4B axis in microglia.
Eur J Pharmacol. 891:1737192021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Aghdam AM, Shahabi P, Karimi-Sales E,
Ghiasi R, Sadigh-Eteghad S, Mahmoudi J and Alipour MR: Swimming
exercise induced reversed expression of miR-96 and its target gene
NaV1.3 in diabetic peripheral neuropathy in rats. Chin J Physiol.
61:124–129. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yan J, Yu H, Shen J, Han C, Li C, Shen X
and Li B: Early over-expressing of microRNA-145 effectively
precludes the development of neuropathic mechanical hyperalgesia
via suppressing Nav1.8 in diabetic rats. Pain Physician.
23:E673–E686. 2020.PubMed/NCBI
|
|
87
|
Wu Y, Gu Y and Shi B: miR-590-3p
Alleviates diabetic peripheral neuropathic pain by targeting RAP1A
and suppressing infiltration by the T cells. Acta Biochim Pol.
67:587–593. 2020.PubMed/NCBI
|
|
88
|
Zhang T, Wang L and Chen L: Alleviative
effect of microRNA-497 on diabetic neuropathic pain in rats in
relation to decreased USP15. Cell Biol Toxicol. 39:1–16. 2023.
View Article : Google Scholar
|
|
89
|
Guo Y, Zeng J, Zhuang Y, Jiang C and Xie
W: MiR-503-5p alleviates peripheral neuropathy-induced neuropathic
pain in T2DM mice by regulating SEPT9 to inhibit astrocyte
activation. Sci Rep. 14:143612024. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kasimu A, Apizi X, Talifujiang D, Ma X,
Fang L and Zhou X: miR-125a-5p in astrocytes attenuates peripheral
neuropathy in type 2 diabetic mice through targeting TRAF6.
Endocrinol Diabetes Nutr (Engl Ed). 69:43–51. 2022.PubMed/NCBI
|
|
91
|
Ashjari D, Karamali N, Rajabinejad M,
Hassani SS, Afshar Hezarkhani L, Afshari D, Gorgin Karaji A, Salari
F and Rezaiemanesh A: The axis of long non-coding RNA
MALAT1/miR-1-3p/CXCR4 is dysregulated in patients with diabetic
neuropathy. Heliyon. 8:e091782022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li YB, Wu Q, Liu J, Fan YZ, Yu KF and Cai
Y: miR-199a-3p is involved in the pathogenesis and progression of
diabetic neuropathy through downregulation of SerpinE2. Mol Med
Rep. 16:2417–2424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wu X, Wang X, Yin Y, Zhu L, Zhang F and
Yang J: Investigation of the role of miR-221 in diabetic peripheral
neuropathy and related molecular mechanisms. Adv Clin Exp Med.
30:623–632. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chang LL, Wang HC, Tseng KY, Su MP, Wang
JY, Chuang YT, Wang YH and Cheng KI: Upregulation of miR-133a-3p in
the sciatic nerve contributes to neuropathic pain development. Mol
Neurobiol. 57:3931–3942. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chen J, Li C, Liu W, Yan B, Hu X and Yang
F: miRNA-155 silencing reduces sciatic nerve injury in diabetic
peripheral neuropathy. J Mol Endocrinol. 63:227–238. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chattopadhyay M, Zhou Z, Hao S, Mata M and
Fink DJ: Reduction of voltage gated sodium channel protein in DRG
by vector mediated miRNA reduces pain in rats with painful diabetic
neuropathy. Mol Pain. 8:172012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Haque S and Harries LW: Circular RNAs
(circRNAs) in health and disease. Genes (Basel). 8:3532017.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang HH, Zhang Y, Wang X, Yang P, Zhang
BY, Hu S, Xu GY and Hu J: Circular RNA profile in diabetic
peripheral neuropathy: Analysis of coexpression networks of
circular RNAs and mRNAs. Epigenomics. 12:843–857. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhang SB, Lin SY, Liu M, Liu CC, Ding HH,
Sun Y, Ma C, Guo RX, Lv YY, Wu SL, et al: CircAnks1a in the spinal
cord regulates hypersensitivity in a rodent model of neuropathic
pain. Nat Commun. 10:41192019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang L, Luo T, Bao Z, Li Y and Bu W:
Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic
rats. Biochem Biophys Res Commun. 505:644–650. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zhang Y, Ma H, Bai Y, Hou X, Yang Y, Wang
G and Li Y: Chronic neuropathic pain and comorbid depression
syndrome: From neural circuit mechanisms to treatment. ACS Chem
Neurosci. 15:2432–2444. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Song Q, Wei A, Xu H, Gu Y, Jiang Y, Dong
N, Zheng C, Wang Q, Gao M, Sun S, et al: An ACC-VTA-ACC
positive-feedback loop mediates the persistence of neuropathic pain
and emotional consequences. Nat Neurosci. 27:272–285. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Huang J, Gadotti VM, Chen L, Souza IA,
Huang S, Wang D, Ramakrishnan C, Deisseroth K, Zhang Z and Zamponi
GW: A neuronal circuit for activating descending modulation of
neuropathic pain. Nat Neurosci. 22:1659–1668. 2019. View Article : Google Scholar : PubMed/NCBI
|















