Introduction
Glioma is the most common primary malignant brain
tumor in adults, and the prognosis and survival rate for patients
with glioma are still extremely poor despite major therapeutic
improvements (1). Particularly,
glioblastoma multiforme is the most malignant form of glioma
according to the current WHO classification of brain tumors
(2,3). Exploring the molecular mechanism
involved in glioma tumorigenesis and progression has become an
urgent demand for developing novel therapeutic strategies for
glioma. Recent studies have reported the alteration of miRNAs in
brain cancer cells, indicating their roles as oncogenes or tumor
suppressors (4–6).
microRNAs (miRNAs) comprise a specialized subset of
small cytoplasmic noncoding RNAs between 19 and 24 nucleotides in
length. It was found that miRNAs play an important role in
regulating various biological processes such as cell proliferation,
differentiation, apoptosis, the cell cycle, migration, invasion,
neuron development and tumorigenesis (4–6).
miRNAs can function as tumor suppressors or oncogenes, depending on
the target genes (7) and mainly
exert their regulatory effects at the post-transcriptional level by
targeting complementary or partly complementary mRNAs and inducing
mRNA cleavage or translation repression (8). microRNA-155 (miR-155) is one of the
miRNAs most frequently involved in cancers, and was initially
implicated in the oncogenesis of hematopoietic malignancies based
on the finding that BIC/miR-155 expression is upregulated in B-cell
lymphomas and chronic lymphocytic leukemia (9). miR-155 was also found to be
overexpressed in various solid tumors, including lung, colon,
breast, cervical, and thyroid cancers (10–13).
Of the mammalian Forkhead transcription factors,
FOXO3a is one member that has emerged as a versatile target for a
number of disorders, including various tumors. Previously, it has
been reported that FOXO3a regulates oxidative detoxification, DNA
damage response and stimulates DNA repair pathways (14,15).
With respect to apoptotic death, Foxo3a has been shown to modulate
a ligand activating a Fas-mediated death pathway (16) and to induce tumor necrosis
factor-related apoptosis-induced ligand (TRAIL) and the BH3-only
proteins Noxa and Bim (17,18). FOXO3a also can modulate growth
arrest and DNA damage response protein 45 (Gadd45), influence cell
cycle regulation (19,20) and induce the repression of
anti-apoptotic molecules FLIP and BCL-XL (21). Shiota et al demonstrated that
FOXO3a may repress cancer cell aggressiveness through negative
regulation of Twist/YB-1 signaling, in addition to its positive
effects on E-cadherin in the progression of urothelial cancer
(22).
Although miR-155 has been found to be upregulated in
glioma, its role has not yet been defined in glioma tumorigenesis.
Based on the prediction of target genes of miR-155, we hypothesized
that there is a significant association between FOXO3a and miR-155.
In the present study, we investigated the effect of miR-155 on the
proliferation and invasion of glioma cells using loss-of-function
and recovery experiments. Furthermore, we aimed to determine
whether miR-155 could directly bind to FOXO3a. The results suggest
that miR-155 serves as an oncomiR and a regulator of FOXO3a
expression in glioma.
Materials and methods
Cell culture and human tissue
samples
The glioma cell line U251 was obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). U87,
U373, SHG44 and T98 cells were gifts from the Neuroscience
Institute of Soochow University. The normal human astrocyte 1800
cell line was obtained from the American Type Culture Collection
(Manassas, VA, USA). All cells were cultured at 37°C in a
humidified atmosphere containing 5% CO2 and maintained
in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Paisley, UK)
supplemented with 10% heat inactivated fetal bovine serum (FBS;
Gibco, USA), 100 μg/ml streptomycin and 1 U/ml penicillin.
Fresh glioma specimens were obtained with written
informed consent from 7 patients with glioma who underwent surgery
at the First Affiliated Hospital of Soochow University. None of the
patients had received radiotherapy or chemotherapy prior to
surgery. Two normal brain tissue samples were obtained from
patients who underwent surgery for cerebral trauma. The resected
tissue samples were immediately snap frozen in liquid nitrogen
until use. All human materials were used in accordance with the
policies of the local institutional review boards.
Real-time PCR
Total RNA was extracted with TRIzol (Invitrogen,
Oslo, Norway). For mature microRNA expression analysis, miR-155 was
detected using an All-in-One™ miRNA qRT-PCR Detection kit
(GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s
instructions. For mRNA, total RNA isolated from glioma samples and
cells were subsequently reverse transcribed to cDNA using
All-in-One First-Strand cDNA Synthesis kit (GeneCopoeia). The
synthesized cDNA was then used to amplify gene regions by
quantitative PCR using the All-in-One qPCR mix (GeneCopoeia catalog
no. AOPR-0200). The U6 small nuclear RNA and GAPDH mRNA were used
as internal controls for miR-155 and FOXO3a mRNA, respectively.
Relative expression was calculated using the ΔΔCt method. The
reactions were placed in a 96-well plate (ABI) using the 7500
Real-Time PCR system (Applied Biosystems).
Western blot analysis
Protein of the treated cell lines was extracted by
mammalian protein extraction reagent (Pierce, USA) supplemented
with protease inhibitors and phosphatase inhibitor cocktail (Sigma,
USA). Protein (25 μg) samples were separated on a 10% SDS-PAGE gel
and then transferred to PVDF membranes. After blocking with 1% BSA
(Amresco, USA), the membranes were incubated with antibodies
against FOXO3a (75D; Cell Signaling, Danvers, MA, USA) diluted
1:1000 or β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) diluted 1:2000 overnight at 4°C; the membrane was washed and
then incubated with horseradish peroxidase (HRP)-conjugated goat
anti-rabbit (Dako, Glostrup, Denmark) or goat anti-mouse (Santa
Cruz Biotechnology, Inc.) secondary antibody diluted 1:2000 for 2
h. The membrane was washed again, and the antigen-antibody reaction
was visualized using an ECL detection system. All experiments were
repeated in triplicate.
miRNA transfection
miR-155 mimics, miR-155 inhibitor and the respective
negative control (miR-NC and anti-NC) were designed and synthesized
by GenePharma (Shanghai, China). The miR-155 inhibitor is
single-stranded RNA molecules, which can specifically knock down
endogenous miR-155. Cells were transfected using Entranster-R
(Engreen Biosystem Co., Ltd., China). Transfection complexes were
prepared according to the manufacturer’s instructions and added
directly to the cells in serum medium. miR-155 mimics and miR-NC
were transfected to U251 cells, which expressed low levels of
miR-155; miR-155 inhibitor (anti-miR-155), inhibitor NC (anti-NC)
were transfected to U87 cells, which expressed high levels of
miR-155.
Cell proliferation
Cell counts were determined using Cell Counting
Kit-8 (Dojindo, Japan). Three thousand cells per well were seeded
in a 96-well plate and incubated for 24 h, and the cells were
transfected with miR-155 mimics, miR-155 inhibitor or NC at the
final concentration of 50 nM/μl. Ten microliters of Cell Counting
Kit-8 was added to 100 μl cell culture media and incubated for 2 h
in CO2 incubator at 12, 24, 48 and 72 h after
transfection. Absorbance was measured at 450 nm. Three independent
experiments were performed.
Cell cycle analysis
Cell cycle distribution was assessed using a flow
cytometer (FC500 Flow Cytometer; Beckman Coulter, USA). According
to the manual from the cell cycle analysis kit (Beyotime, China),
cells were allowed to inoculate and adhere for 24 h in 6-well
plates. Then miR-155 mimics, miR-155 inhibitor and NC were
transfected into cells of the respective well, and cells were
incubated for a further period of 24 h, then trypsinized, washed
three times in cold phosphate-buffered saline (PBS), fixed in 70%
absolute ice-cold ethanol, kept at 4°C overnight and stained in a
mixture of propidium (PI) RNase A; the stained cells were analyzed
using flow cytometry. This experiment was repeated three times.
Cell apoptosis assay
Cells were transfected with miR-155, anti-miR-155,
or NC at 50% confluence. After 48 h, the adherent cells were
harvested by trypsinization and were washed with PBS once and
resuspended in 500 μl of 1X binding buffer (Annexin V/FITC kit;
Sigma). Then, 10 μl Annexin V/FITC and 5 μl propidium iodide were
added into the binding buffer, and the tubes were incubated at room
temperature for exactly 10 min in the dark. The fluorescence of the
cells was immediately determined with a flow cytometer.
Monolayer wound healing assay
Migratory ability was determined using a wound
healing assay. Cells were grown in 10% FBS medium on 6-well plates.
After the cells reached sub-confluence, the cells were wounded by
scraping the monolayer with a 200-μl pipette tip and grown in 1%
FBS medium. The floating cells were removed by gentle washes with
culture medium. The healing process was dynamically examined and
was recorded with a digital camera at 24 h after the wound was
created.
Transwell invasion assay
For the Transwell assay, 2×104
transfected cells were plated into 24-well Boyden chambers (Corning
Costar, Cambridge, MA, USA), with an 8-μm pore polycarbonate
membrane, which was coated with 30 μg of Matrigel (BD Biosciences,
San Jose, CA, USA). Cells were in the upper chamber with 200 μl of
serum-free medium, and medium containing 20% FBS was added to the
lower chamber to serve as a chemoattractant. After 36 h, the cells
were washed 3 times with PBS. Non-invasive cells were removed from
the upper well by cotton swabs, and the invasive cells were then
fixed with paraformaldehyde for 15 min, air-dried, and stained with
0.1% crystal violet for 15 min. The cells were recorded with a
digital camera.
Luciferase activity assay
DNA fragments from 3′-UTR of FOXO3a that host the
predicted complementary sites of miR-155 were cloned to downstream
of the Renilla luciferase reporter gene in psiCHECK2 dual
luciferase reporter plasmid (Promega, Madison, WI, USA). We
amplified a 2192-bp FOXO3a 3′-UTR region containing four tandem
repeats of miR-155 response element, which includes the artificial
XhoI and NotI enzyme restriction sites with forward
primer 5′-TGTTGTTCTTGTG TTTGTTTTCC-3′ and reverse primer
5′-ATTCTCCTGATC TGTTTTGTGCT-3′. The amplified fragments were then
cleaved with the XhoI and NotI enzymes (New England
BioLabs, Ipswich, MA, USA) as well as psiCHECK2 vector (Promega),
and the above-prepared fragment and psiCHECK2 vector were then
ligated by T4 DNA ligase (New England BioLabs). The recombined
vector was named psi-FOXO3a.
U251 or U87 cells were seeded at 1×105
cells per well in 24-well plates. At 24 h after the plating, cells
were transfected by Lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA) according to the manufacturer’s instructions. In each well,
800 ng psi-FOXO3a vectors were co-transfected with 50 pmol miR-155
mimics, miR-155 inhibitors or NC accordingly. There were four
replicates for each group, and the experiment was repeated at least
three times. At 24 h after transfection, cells were harvested by
passive adding of 100 μl buffer. Renilla luciferase
activities in the cell lysate were measured with the
Dual-Luciferase Reporter assay system (Promega) in TD-20/20
luminometer (Turner Biosystems, Sunnyvale, CA, USA) and were
normalized to the firefly luciferase activities.
Statistical analysis
Data shown in the graphs represent the mean values ±
SD of three independent experiments performed. The difference among
groups was determined by ANOVA analysis and comparison between two
groups was analyzed by the Student’s t-test using GraphPad Prism
software version 4.0 (GraphPad Software, Inc., San Diego, CA, USA).
A value of P<0.05 was considered statistically significant.
Results
Expression of FOXO3a in glioma cell lines
and human glioma tissues
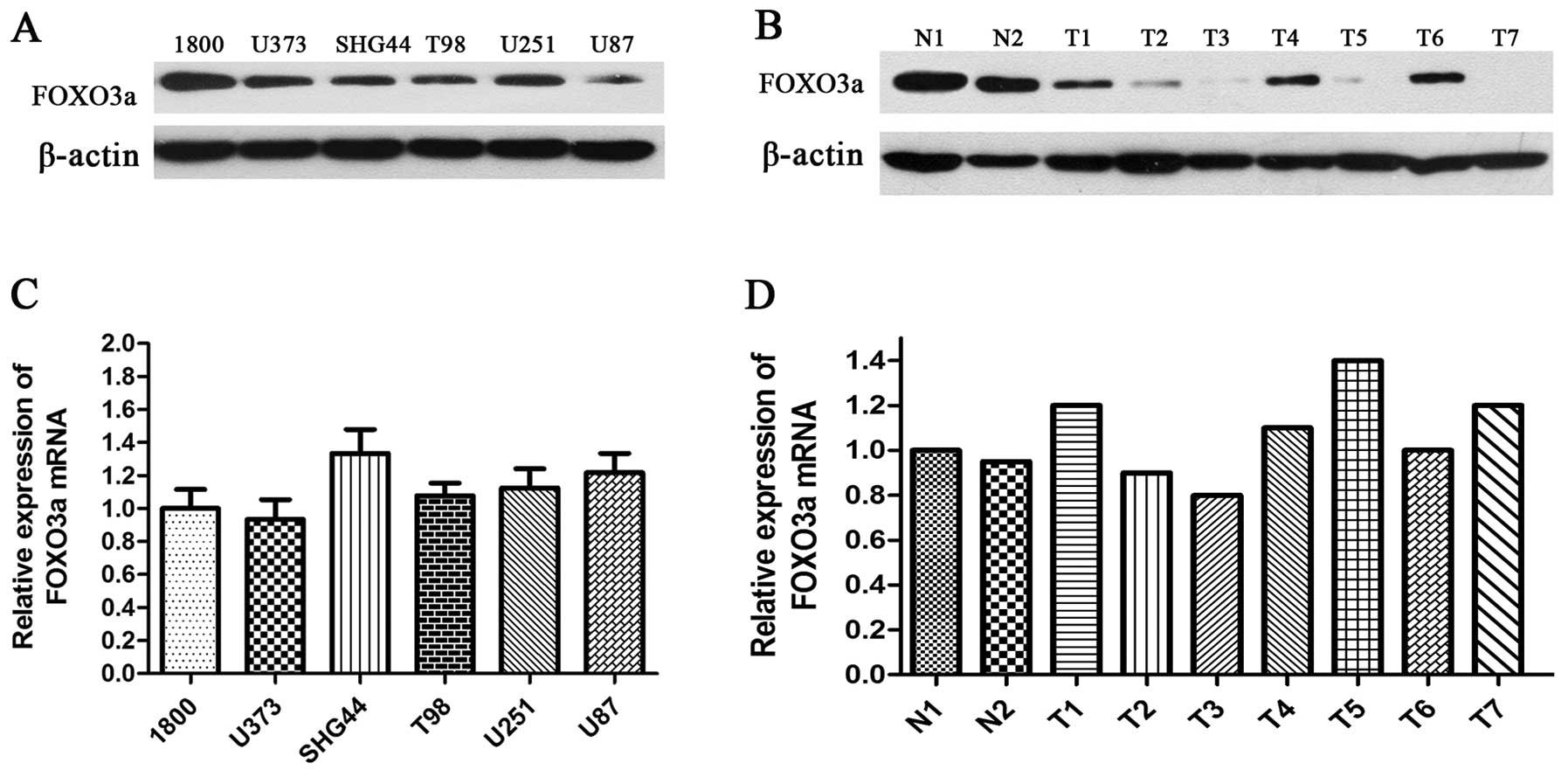
To explore the role of FOXO3a in glioma, we used
western blotting to detect the expression level of FOXO3a in glioma
cell lines, human glioma and normal brain tissues. The findings
showed that five carcinoma cell lines (U251, U87, U373, T98 and
SHG44) presented lower expression of FOXO3a protein when compared
with the normal astrocyte 1800 cell line (Fig. 1A). Among all glioma cell lines, the
highest expression of FOXO3a was detected in U251 cells, while the
lowest expression was observed in U87 cells. Moreover, FOXO3a
levels in glioma tissue were significantly lower than those in
normal brain tissues (Fig. 1B).
However, no significant difference in FOXO3a mRNA was observed
between glioma cell lines, and between human glioma tissues,
respectively (Fig. 1C and D).
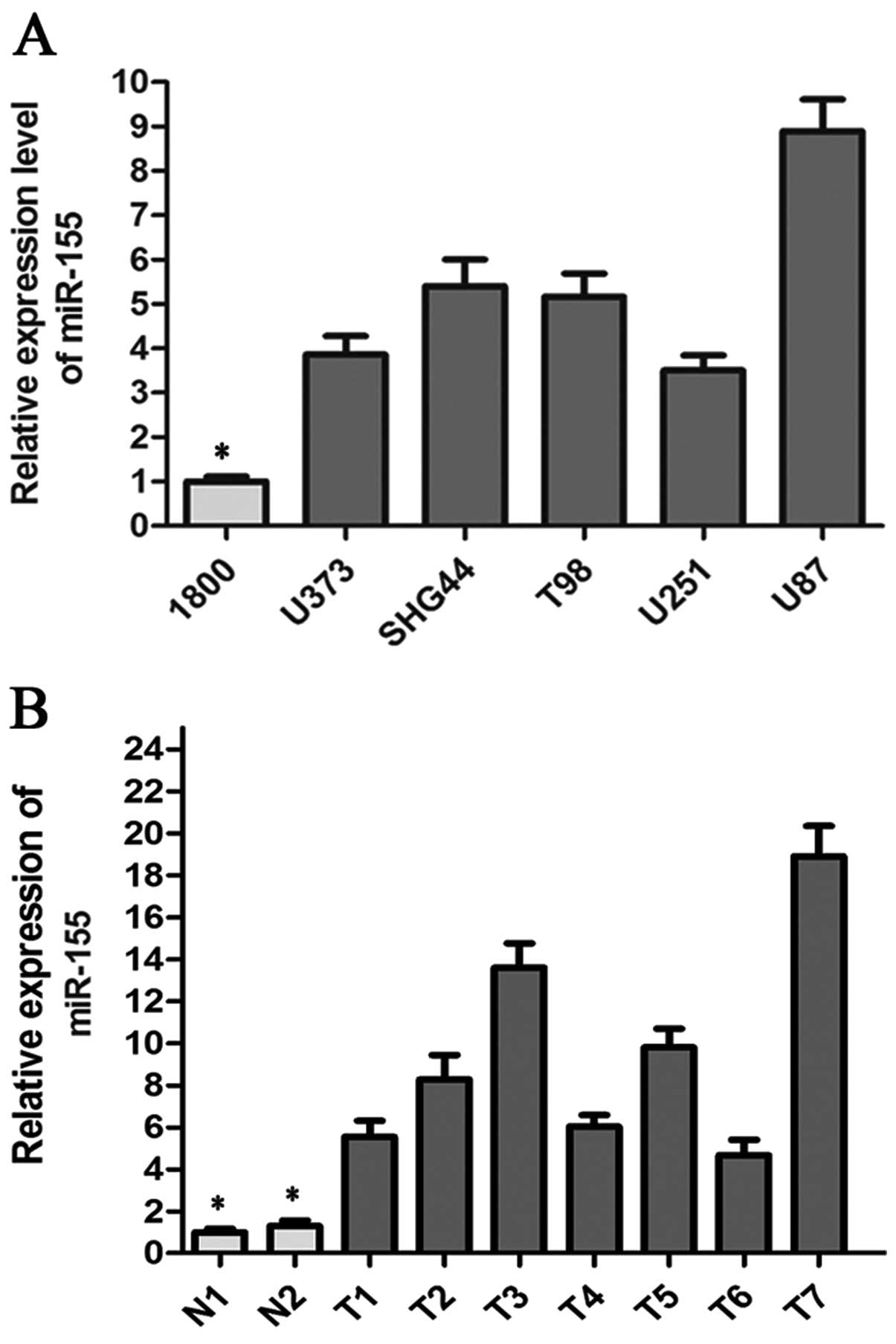
miR-155 is upregulated in glioma cell
lines and the glioma samples
To determine whether miR-155 plays a role in glioma
tumorigenesis, we conducted real-time quantitative PCR to quantify
mature miR-155 in five glioma cell lines (U251, U87, U373, T98 and
SHG44) and seven glioma tissue samples, using the normal human
astrocyte 1800 cell line and two normal brain tissue samples as
corresponding controls. As shown in Fig. 2A, miR-155 expression was notably
upregulated in five glioma cell lines, when compared to that in the
normal astrocyte 1800 cell line. Among all glioma cell lines, the
U87 cell line expressed the highest level of miR-155 (9-fold
compared to the normal control) and U251 cell line expressed the
lowest level of miR-155 (3.5-fold compared to the normal control)
(Fig. 2A). Higher expression of
miR-155 was observed in all glioma tissue samples (Fig. 2B).
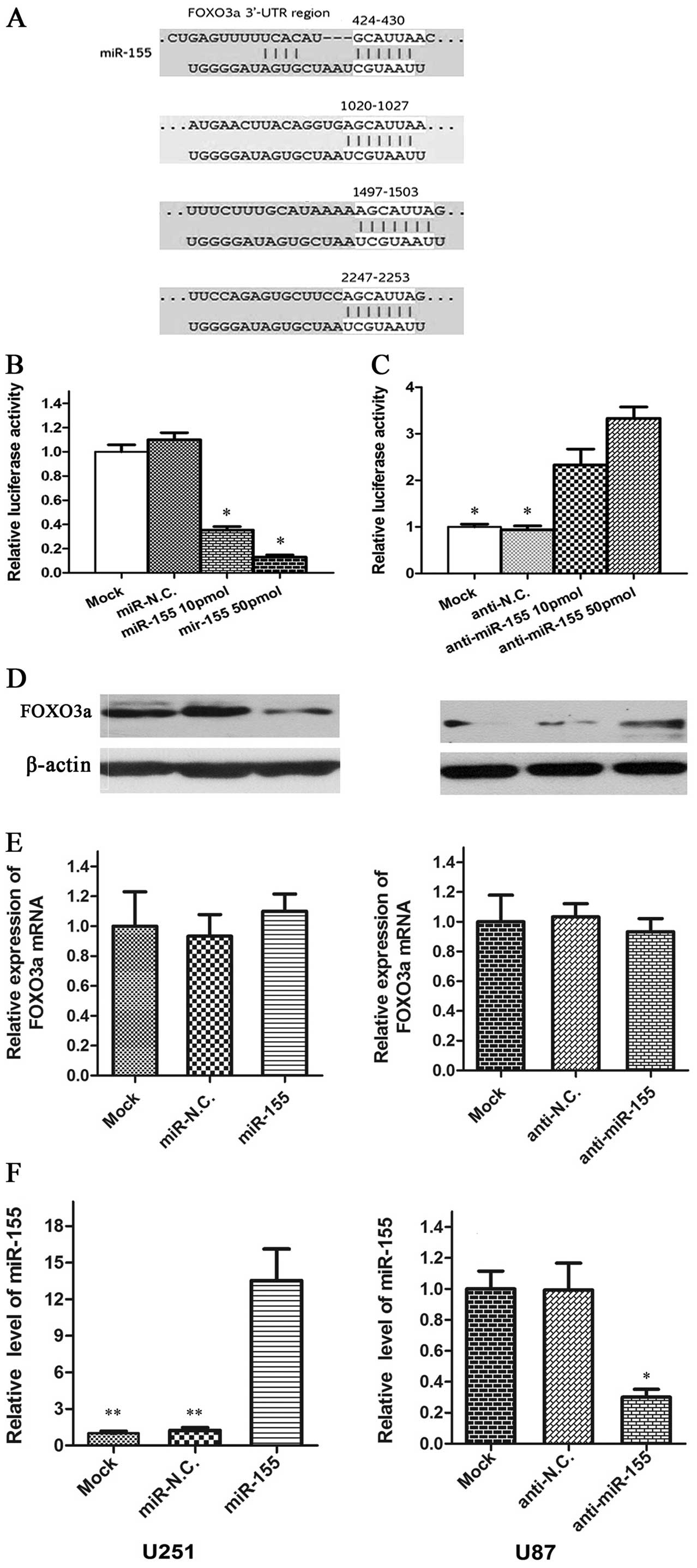
FOXO3a is a direct target of miR-155
TargetScan prediction in silico and previous
studies indicated that FOXO3a is the theoretical target gene of
miR-155 (Fig. 3A). To test this
possibility, fragments of 3′-UTR of human FOXO3a containing miR-155
complementary sites were cloned into psiCHECK2 dual luciferase
reporter plasmid. When the psi-FOXO3a vector was co-transfected
with miR-155, the luciferase activity of the vector was
dramatically decreased (P<0.05) compared with those
co-transfected with miR-NC and Mock in U251 cells (Fig. 3B). As expected, the luciferase
levels of psi-FOXO3a-3′-UTR in U87 cell lines were restored after
transfection with the miR-155 inhibitor (Fig. 3C).
By performing western blot analysis, a pronounced
reduction in FOXO3a protein level was noted in the U251 cells in
which miR-155 is overexpressed and an increase in U87 cells in
which miR-155 is downregulated (Fig.
3D). However, no significant difference in FOXO3a mRNA was
observed between cells overexpressing/underexpressing miR-155 and
those with NC (Fig. 3E and F). This
raised the possibility that miR-155 suppresses FOXO3a protein
synthesis by a post-transcriptional repression mechanism, via its
3′-UTR complementary sites.
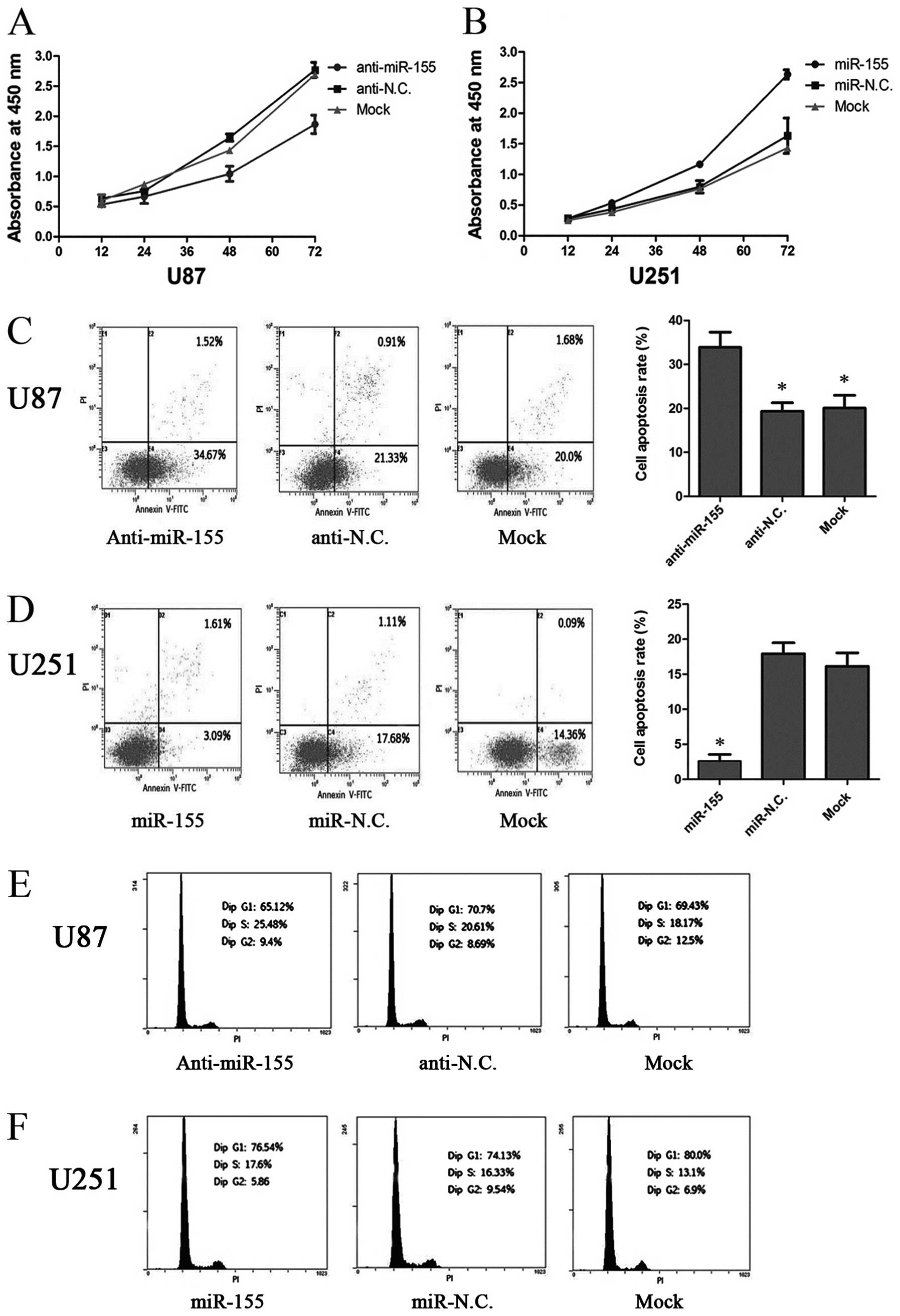
miR-155 increases cell proliferation by
inhibiting apoptosis in glioma cells
In order to explore the functional role of miR-155
on the growth of glioma cells, we performed cellular proliferation
assays (CCK-8 assay) in U87 and U251 cells and found that cell
proliferation was dramatically decreased in U87 cells after
transfection with the miR-155 inhibitor, while proliferation was
increased in U251 cells transfected with miR-155 mimics (P<0.05)
(Fig. 4A and B). Then, we carried
out cell apoptosis and cell cycle assays to examine whether the
reduction in cell proliferation was due to increased apoptosis or
attenuated cell cycle induced by downregulation of miR-155. The U87
cells transfected with the miR-155 inhibitor were found to exhibit
an enhanced apoptosis rate, compared to the anti-NC and Mock
groups, while U251 cells transfected with miR-155 mimics exhibited
the opposite results (Fig. 4C and
D), while the alteration of miR-155 expression was ineffective
on the cell cycle distribution of glioma cells (Fig. 4E and F). These results suggest that
miR-155 increases the cell proliferative ability by inhibiting
apoptosis in glioma cells.
miR-155 promotes the migration and
invasion of glioma cells
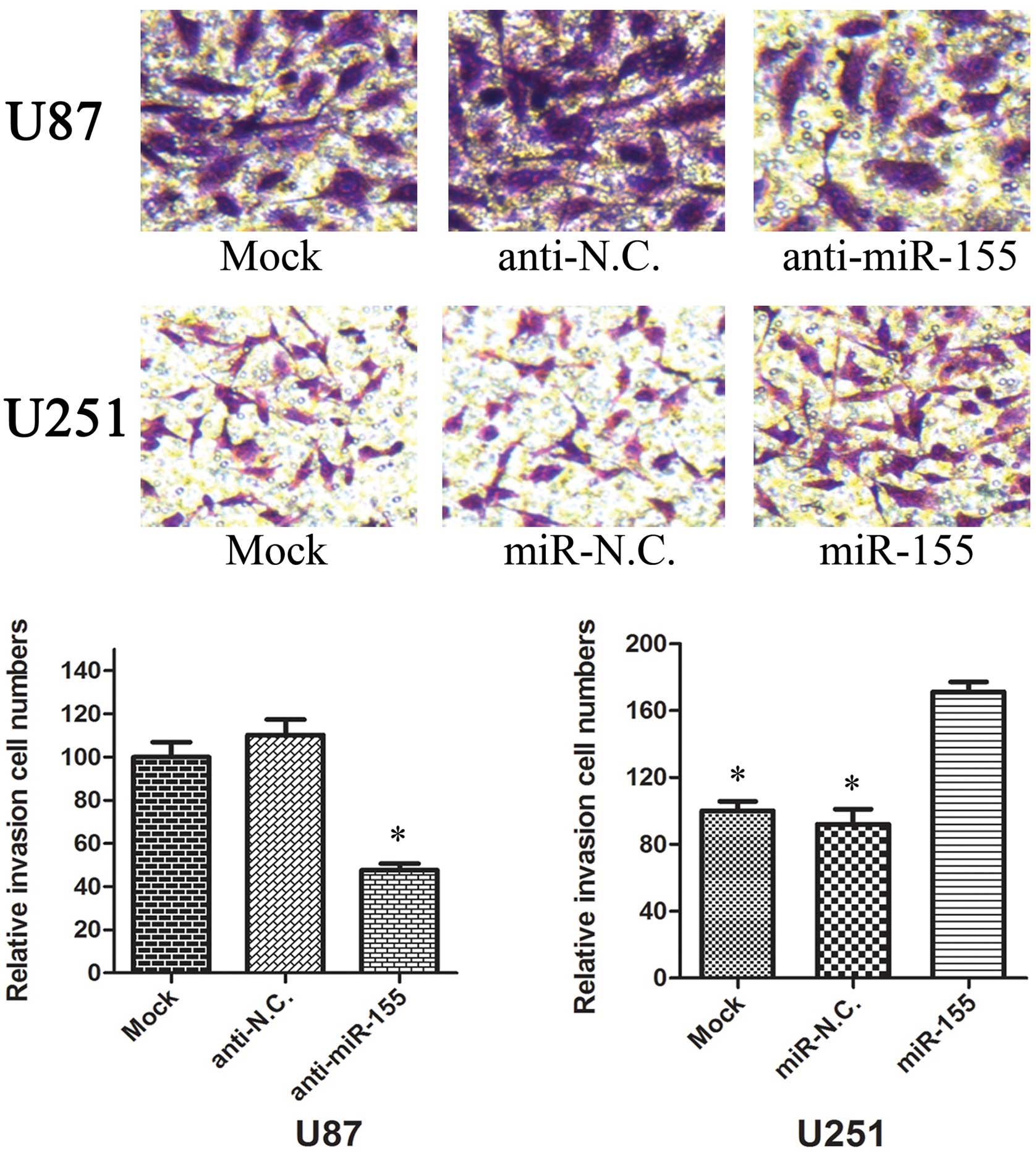
To investigate whether miR-155 has a direct
functional role in facilitating glioma cell migration and invasion,
we evaluated cancer cell invasion through Transwell assay and
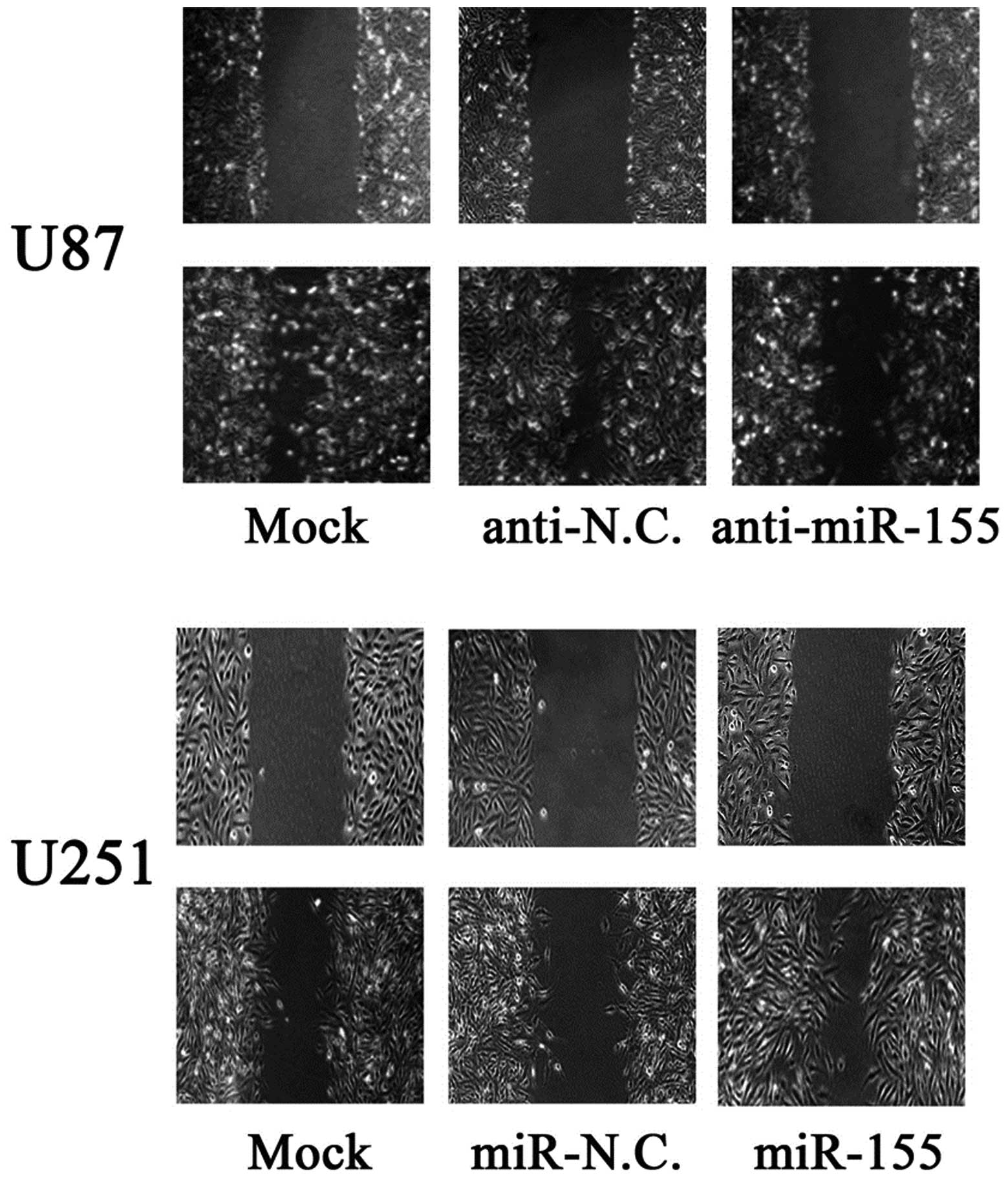
assessed migration through a wound healing assay. As shown in
Fig. 5, inhibition of miR-155
impeded the migration of U87 cells by ~50% when compared with the
control. Similarly, invasion of U87 cells was reduced by 59%
following inhibition of miR-155 (Fig.
6). Conversely, transfection of U251 cells with miR-155 mimics
promoted cell migration and invasion ability by 2-fold (Fig. 5 and 6). These data suggest that miR-155 is an
onco-miRNA that can promote cell migration and invasion in glioma
cells.
Discussion
miR-155 has emerged as an essential regulator of
cellular physiology, and deregulation of miR-155 is implicated in a
wide range of diseases, including pituitary adenoma, multiple
malignant tumors, myelodysplastic syndrome and Down syndrome
(23–26). miR-155 has been extensively
investigated in immunology and lymphoma; however, it is only
evident that miR-155 expression is increased in glioma, and to date
the detailed function of miR-155 remains elusive. In the present
study, we also found that miR-155 was upregulated in glioma cell
lines and all glioma tissue samples, and we transfected the miR-155
inhibitor and miR-155 mimics into U87 and U251 cells, repectively,
to evaluate glioma cell proliferation, the cell cycle, apoptosis,
migration and invasion. The results suggest that miR-155 may not
only enhance cell proliferative ability by inhibiting apoptosis but
also may promote cell migration and invasion in glioma cells.
To further elucidate the molecular mechanisms and
improve our understanding of the regulatory function of miR-155 in
glioma, we first analyzed the expression of FOXO3a, which was
predicated as a target of miR-155, and found that expression of
FOXO3a protein was markedly attenuated in glioma cells and human
glioma tissues. It was reported that inactivation of FOXO3a in
glioma cells led to the downregulation of Bim, a pro-apoptotic
protein (20). Hu and colleagues
demonstrated that expression of FOXO3a exerts an inhibitory effect
on tumorigenesis and cell growth in breast cancer cells (27). In cancer cells, suppression of
FOXO3a was believed to be due mainly to the activation of multiple
onco-kinases such as Akt, SGK and IKK. However, the dysregulation
of FOXO3a can be caused by various mechanisms, including miRNA
regulation.
Subsequently, we used two different methods to
determine whether FOXO3a is a bonafide target of miR-155.
Overexpression of miR-155 caused the reduction in FOXO3a protein
albeit there was no difference in FOXO3a mRNA expression between
miR-155-overexpressing and negative control cells. These results
are consistent with previous data, which indicated that
miR-155-induced silencing appears to occur only through
translational repression without affecting mRNA levels (28). Furthermore, the luciferase activity
assay demonstrated that downregulation of FOXO3a was mediated by
miR-155, which may target 3′-UTR of FOXO3a. Thus, our findings
support the hypothesis that the effect of miR-155 on proliferation
and invasion of glioma cells may be attributed to the
downregulation of FOXO3a, via directly targeting the
FOXO3a-3′-UTR.
However, an opposite trend in the expression of
miR-155 has been observed in other types of cancer. For example,
miR-155 was found to be downregulated in melanoma when compared
with non-cancerous tissue and could mediate melanoma growth
inhibition via SKI gene silencing (29). As exemplified by miR-10b, it was
found to be overexpressed in glioma and induced glioma cell
invasion by modulating tumor invasion factors MMP-14 and uPAR
expression via the direct target HOXD10 (30). However, Moriarty et al found
that miR-10b was downregulated and inhibited Tiam1-mediated Rac
activation to suppress migration and invasion of breast carcinoma
(31). In addition, overexpression
of miR-30 inhibited apoptosis via suppression of p53 expression in
cardiomyocytes (32), while
upregulation of miR-30 induced apoptosis by targeting Ubc9 in
breast cancer cells (33),
suggesting that a single miRNA may have several distinct functions
in different cell types, which likely depends on the availability
of specific targets or downstream effectors. Therefore, it is
extremely important to validate targets of miR-155 by further
functional assays.
In summary, we demonstrated that miR-155 is markedly
upregulated in human glioma, and is able to downregulate FOXO3a
expression to enhance the proliferative and invasive ability of
glioma cells, suggesting that silencing of miR-155 expression may
be a novel strategy for the treatment of human glioma.
Acknowledgements
This study was supported in part by grants from the
Program for New Century Excellent Talents in University
(NCET-09-0165 to H.-T. Zhang), Jiangsu Province’s Key Provincial
Talents Program (RC2011106 to J. Zhao), ‘333’ Project of Jiangsu
Province Government (to H.-T. Zhang), Soochow Scholar Project of
Soochow University (to H.-T. Zhang), and Suzhou Key Laboratory for
Molecular Cancer Genetics (SZS201209 to H.-T. Zhang).
References
|
1
|
Behin A, Hoang-Xuan K, Carpentier AF and
Delattre JY: Primary brain tumours in adults. Lancet. 361:323–331.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou H, Miki R, Eeva M, Fike FM, Seligson
D, Yang L, Yoshimura A, Teitell MA, Jamieson CA and Cacalano NA:
Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to
ionizing radiation in glioblastoma multiforme. Clin Cancer Res.
13:2344–2353. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohgaki H: Genetic pathways to
glioblastomas. Neuropathology. 25:1–7. 2005. View Article : Google Scholar
|
|
4
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36.
2003. View Article : Google Scholar
|
|
6
|
D’Urso PI, D’Urso OF, Storelli C, Mallardo
M, Gianfreda CD, Montinaro A, Cimmino A, Pietro C and Marsigliante
S: miR-155 is up-regulated in primary and secondary glioblastoma
and promotes tumour growth by inhibiting GABA receptors. Int J
Oncol. 41:228–234. 2012.PubMed/NCBI
|
|
7
|
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT,
Goan YG and Lu PJ: MicroRNA-330 acts as tumor suppressor and
induces apoptosis of prostate cancer cells through E2F1-mediated
suppression of Akt phosphorylation. Oncogene. 28:3360–3370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yekta S, Shih IH and Bartel DP:
MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Faraoni I, Antonetti FR, Cardone J and
Bonmassar E: miR-155 gene: a typical multifunctional microRNA.
Biochim Biophys Acta. 1792:497–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin
GA, Liu CG, Croce CM and Harris CC: Unique microRNA molecular
profiles in lung cancer diagnosis and prognosis. Cancer Cell.
9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yip L, Kelly L, Shuai Y, Armstrong MJ,
Nikiforov YE, Carty SE and Nikiforova MN: MicroRNA signature
distinguishes the degree of aggressiveness of papillary thyroid
carcinoma. Ann Surg Oncol. 18:2035–2041. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I,
Calin GA, Querzoli P, Negrini M and Croce CM: MicroRNA gene
expression deregulation in human breast cancer. Cancer Res.
65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tran H, Brunet A, Grenier JM, Datta SR,
Fornace AJ Jr, DiStefano PS, Chiang LW and Greenberg ME: DNA repair
pathway stimulated by the forkhead transcription factor FOXO3a
through the Gadd45 protein. Science. 296:530–534. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsai WB, Chung YM, Takahashi Y, Xu Z and
Hu MC: Functional interaction between FOXO3a and ATM regulates DNA
damage response. Nat Cell Biol. 10:460–467. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barthelemy C, Henderson CE and Pettmann B:
Foxo3a induces motoneuron death through the Fas pathway in
cooperation with JNK. BMC Neurosci. 5:482004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Obexer P, Geiger K, Ambros PF, Meister B
and Ausserlechner MJ: FKHRL1-mediated expression of Noxa and Bim
induces apoptosis via the mitochondria in neuroblastoma cells. Cell
Death Differ. 14:534–547. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Delpuech O, Griffiths B, East P, Essafi A,
Lam EW, Burgering B, Downward J and Schulze A: Induction of Mxi1-SR
alpha by FOXO3a contributes to repression of Myc-dependent gene
expression. Mol Cell Biol. 27:4917–4930. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Furukawa-Hibi Y, Yoshida-Araki K, Ohta T,
Ikeda K and Motoyama N: FOXO forkhead transcription factors induce
G(2)-M checkpoint in response to oxidative stress. J Biol Chem.
277:26729–26732. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo
P, Hu LS, Anderson MJ, Arden KC, Blenis J and Greenberg ME: Akt
promotes cell survival by phosphorylating and inhibiting a Forkhead
transcription factor. Cell. 96:857–868. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang TT, Dowbenko D, Jackson A, Toney L,
Lewin DA, Dent AL and Lasky LA: The forkhead transcription factor
AFX activates apoptosis by induction of the BCL-6 transcriptional
repressor. J Biol Chem. 277:14255–14265. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shiota M, Song Y, Yokomizo A, Kiyoshima K,
Tada Y, Uchino H, Uchiumi T, Inokuchi J, Oda Y, Kuroiwa K,
Tatsugami K and Naito S: Foxo3a suppression of urothelial cancer
invasiveness through Twist1, Y-box-binding protein 1, and
E-cadherin regulation. Clin Cancer Res. 16:5654–5663. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Butz H, Liko I, Czirjak S, Igaz P, Khan
MM, Zivkovic V, Balint K, Korbonits M, Racz K and Patocs A:
Down-regulation of Wee1 kinase by a specific subset of microRNA in
human sporadic pituitary adenomas. J Clin Endocrinol Metab.
95:E181–E191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang
Y, Zhao J, McCrae MA and Zhuang H: Aberrant expression of microRNA
155 may accelerate cell proliferation by targeting sex-determining
region Y box 6 in hepatocellular carcinoma. Cancer. 118:2431–2442.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuhn DE, Nuovo GJ, Terry AV Jr, Martin MM,
Malana GE, Sansom SE, Pleister AP, Beck WD, Head E, Feldman DS and
Elton TS: Chromosome 21-derived microRNAs provide an etiological
basis for aberrant protein expression in human Down syndrome
brains. J Biol Chem. 285:1529–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Santamaria C, Muntion S, Roson B, Blanco
B, Lopez-Villar O, Carrancio S, Sanchez-Guijo FM, Diez-Campelo M,
Alvarez-Fernandez S, Sarasquete ME, de las Rivas J, Gonzalez M, San
Miguel JF and Del Canizo MC: Impaired expression of DICER, DROSHA,
SBDS and some microRNAs in mesenchymal stromal cells from
myelodysplastic syndrome patients. Haematologica. 97:1218–1224.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang
F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R and Hung
MC: IkappaB kinase promotes tumorigenesis through inhibition of
forkhead FOXO3a. Cell. 117:225–237. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong W, He L, Coppola M, Guo J, Esposito
NN, Coppola D and Cheng JQ: MicroRNA-155 regulates cell survival,
growth, and chemosensitivity by targeting FOXO3a in breast cancer.
J Biol Chem. 285:17869–17879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Levati L, Pagani E, Romani S, Castiglia D,
Piccinni E, Covaciu C, Caporaso P, Bondanza S, Antonetti FR,
Bonmassar E, Martelli F, Alvino E and D’Atri S: MicroRNA-155
targets the SKI gene in human melanoma cell lines. Pigment Cell
Melanoma Res. 24:538–550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang
J, Wang X, You Y, Yang Z and Liu N: MicroRNA-10b induces glioma
cell invasion by modulating MMP-14 and uPAR expression via HOXD10.
Brain Res. 1389:9–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moriarty CH, Pursell B and Mercurio AM:
miR-10b targets Tiam1: implications for Rac activation and
carcinoma migration. J Biol Chem. 285:20541–20546. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li J, Donath S, Li Y, Qin D, Prabhakar BS
and Li P: miR-30 regulates mitochondrial fission through targeting
p53 and the dynamin-related protein-1 pathway. PLoS Genet.
6:e10007952010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu F, Deng H, Yao H, Liu Q, Su F and Song
E: Mir-30 reduction maintains self-renewal and inhibits apoptosis
in breast tumor-initiating cells. Oncogene. 29:4194–4204. 2010.
View Article : Google Scholar : PubMed/NCBI
|