Introduction
Hepatocellular carcinoma (HCC), as the most common
type of primary liver cancer, is the sixth most common cancer
globally, with a prevalence of ~600,000 individuals worldwide. HCC
is the second most lethal cancer worldwide and one of the most
rapidly growing cancer types in Asia, particularly in China
(1,2). Reaction of biochemistry and gene
control of apoptosis are important in hepatocarcinogenesis. Failure
of apoptosis further promotes the development of cancer, owing to
tumor occurrence. The majority of HCC cells are markedly resistant
to the stimuli of inducing apoptosis, which can lead to HCC cell
anti-apoptosis (3). Many factors
prevent HCC cells from apoptosis, including anti-apoptotic members
Bcl-2/xL (4), survivin and
cyclooxygenase (COX)-2 (5,6). The abnormal expression of these
factors contributes to resistance to HCC cell apoptosis, resulting
in loss of tumor control and patient death. Therefore,
identification of a new mechanism to convert resistance to
apoptosis and promote apoptosis for the treatment of HCC is
crucial.
TRAIL (TNF-related apoptosis-inducing ligand), also
known as Apo2 ligand (Apo2L), is a member of the TNF family, and
has been shown to induce cell apoptosis in various types of tumor
but not normal cells (7). Five
receptors, including TRAIL-R1/DR4, TRAIL-R2/DR5, TRAIL-R3/DcR1,
TRAIL-R4/DcR2 and TNFR-homologue osteoprotegerin (OPG) have been
identified thus far (8). However,
of these receptors, only TRAIL-R1/DR4 and TRAIL-R2/DR5 induce
apoptosis (8,9). Several tumor cell lines exhibiting
resistance to TRAIL-mediated apoptosis include HCC cell lines
(3,10,11).
However, the mechanism involved in the resistance to TRAIL-induced
apoptosis in HCC cells is mainly connected with activation of the
NF-κB pathway, upregulation of apoptotic inhibitors, such as cFLIP,
IAP and anti-apoptotic molecules, such as certain Bcl-family
members (12–15). TRAIL is involved in inhibition of
cancer. Thus, the upregulation of HCC cell sensitization in
TRAIL-induced apoptosis by mediating the above targets is
significant in HCC treatment.
SNAIL, as a transcriptional repressor, has a
structure of zinc finger (ZF) which plays an important role in
physiological processes such as embryonic development, and in a
wide variety of pathologic processes (16,17).
SNAIL has been found to enhance cancer invasion and metastasis in
various malignancies (16,18,19).
Similarly, SNAIL is crucial to the pathological progression of HCC.
Evidence suggests that SNAIL contributed to tumor progression by
inducing EMT to transform epithelial cells into mesenchymal ones
(18). In addition, a study showed
that SNAIL was induced and accelerated cell activity by repressing
E-cadherin expression and upregulating MMP expression in HCC both
in vitro or in vivo (19). Findings of another study showed that
knockdown of SNAIL reduced proliferation and viability of HCC cells
by increasing the expression of E-cadherin (20). All the aforementioned mechanisms
suggested that SNAIL affects the viability and migration of HCC
cells. Nevertheless, whether SNAIL affects the apoptosis of HCC
cells remains to be determined. Thus, it is imperative to examine
the characteristics of SNAIL with regard to the occurrence and
progression of HCC, particularly in cell apoptosis.
RNA interference (RNAi) has emerged as a new and an
indispensable tool for loss of gene function in eukaryotes in order
for small-interfering RNA (siRNA), a class of synthetic short
double-stranded RNA, to emerges at the appopriate time point
(21,22). siRNA functions by inducing the
silencing of gene by guiding endonucleolytic cleavage of message
RNA (mRNA) or repressing its translation (23,24),
while siRNA cannot maintain long-term silencing of the interfering
gene. Thus, to induce the gene to stable and reversible silencing
for a long period of time, short hairpin RNA (shRNA) was generated
as a tool of the RNAi technique (21). A variety of viral and non-viral
vectors can carry and express shRNA (22). Recently, studies verified successful
construction of the lentivirus-shSNAIL vector, which could be
infected into the HepG2 cell line to silence SNAIL (20). Adenoviral vectors harboring TRAIL
were constructed by our laboratory previously (25), which all provide powerful support
for our study.
In the present study, by silencing SNAIL via the
utilization of RNAi in HCC cells, we investigated the effect of
SNAIL deficiency for HCC cell apoptosis and the sensitization
effect for TRAIL-induced apoptosis by downregulating SNAIL in HCC
cells. The possible mechanism involved was also assessed.
Materials and methods
Cell line and cell culture
The human HCC HepG2, HuH-7 and HEK293 (human
embryonic kidney cells) cell lines were purchased from the American
Type Culture Collection (Manassas, VA, USA). SMMC7721 was purchased
from the Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences, Shanghai, China. The cells were maintained in
a humidified condition that contained 5% CO2 at 37°C and
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS) and 2 mM glutamine, followed by the addition of 100
U/ml penicillin and 100 μg/ml streptomycin.
Construction of lentiviral and adenoviral
vectors
The most effective lentiviral vectors carrying shRNA
were previously screened in our laboratory (20). Adenoviral vectors harboring TRAIL
and lentiviral vectors carrying shBcl-xL, shcIAP2, shSurvivin and
shRaf-1 were previously constructed by our laboratory (20,25).
Construction and purification of the vectors were performed as per
the standard protocol. The titration of recombinant lentiviruses
and adenoviruses was performed using a TCID50 assay on HEK293
cells. Recombinant lentiviral and adenoviral vectors were short for
LV and Ad5, respectively. The cells were subsequently divided into
the Mock, LV-shSNAIL, Ad5.TRAIL and Ad5. TRAIL + LV-shSNAIL
groups.
Quantification by real-time PCR
Infected cells were collected and washed with
phosphate-buffered saline (PBS). Total RNA was extracted from the
cells of the target cells, and cDNA was generated with a
PrimeScript® RT reagent kit (Takara, Chiga, Japan) at a
total volume of 20 μl according to the manufacturer’s instructions.
cDNA was then used in each amplification reaction. Reactions were
performed using SYBR® Premix Ex Taq™ (Takara), under the
following PCR conditions: denaturation at 95°C for 30 sec, followed
by 40 cycles of annealing at 95°C for 5 sec and extension at 60°C
for 30 sec. Each sample was also subjected to melting curve
analysis to confirm amplification specificity. GAPDH was
used as a control housekeeping gene. The expression of SNAIL was
assessed by normalization of the cycle threshold (Ct) of these
genes to that of GAPDH. A Ct value was obtained from each
amplification curve by using the software provided by the
manufacturer (Roche, Mannheim, Germany).
Cell proliferation detection
The analysis of HCC cell viability was determined by
an MTT [3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium
bromide] assay. HCC cells were plated in 96-well microtiter plates
equally according to the density standards of 5×103
cells/well. Each well was provided suitable culture medium with the
condition in 5% CO2 at 37°C until grown to a high
density. Each group of cells was incubated for 24–96 h following
treatment with various viruses. Subsequently, 5 mg/ml MTT was added
to each well and incubated for 4 h under the same condition. The
original culture of each well was abandoned. The crystal substances
produced from the cells were dissolved in 150 μl dimethyl sulfoxide
(DMSO) and agitated for 10 min. Cell viability was assessed as
optical density (OD) at a wavelength of 490 nm for immune
monitoring by the enzyme-linked immunosorbent spot assay (Bio-Rad,
Hercules, CA, USA).
Western blot analysis
Whole-cell lysates were obtained following
centrifugation at 120,000 × g for 10 min of the target cells of all
the groups. Total proteins were separated by electrophoresis on 12%
polyacrylamide and transferred to 0.45 μm nitrocellulose (NC)
membrane. The samples were washed three times using PBS after
Ponceau S Staining kit dyeing. Bands were incubated with primary
antibodies, rabbit polyclonal anti-SNAIL Santa Cruz Biotechnology,
Inc. (Santa Cruz, CA, USA) and anti-TRAIL (Cell Signaling
Technology, Danvers, MA, USA), respectively. Conjugates were
combined with secondary goat anti-rabbit alkaline phosphatase
antibody (Abcam, Cambridge, MA, USA), which was marked with HRP
(horseradish peroxidase). GAPDH detection was performed using
rabbit polyclonal anti-GAPDH and mouse monoclonal anti-GAPDH
(Abcam). The fluorescent compounds were detected by ECL (Pierce,
Rockford, IL, USA). At the same time, primary antibodies, rabbit
polyclonal anti-p53 (Cell Signaling Technology), anti-Bcl-xL (Cell
Signaling Technology), anti-cIAP2 (Abcam, Cambridge, UK),
anti-survivin (Cell Signaling Technology), anti-Raf-1 (Cell
Signaling Technology), respectively, were applied to the membranes
and the process was repeated as above.
Hoechst test
HCC cell lines were placed into 6-well plates and
incubated with recombinant lentiviruses or adenoviruses,
respectively. After 48 h, when the cells were infected, the medium
was washed with PBS twice. The cells were then stained with Hoechst
33258 (25 μg/ml). Hoechst 33258 was excited by 405-nm violet diode.
The percentage of apoptotic cells was analyzed using a fluorescence
microscope.
Statistical analysis
The statistical analyses were performed using the
Student’s t test and one-way ANOVA to determine the significance.
Results were presented as mean ± standard deviation (SD). P<0.05
was considered statistically different.
Results
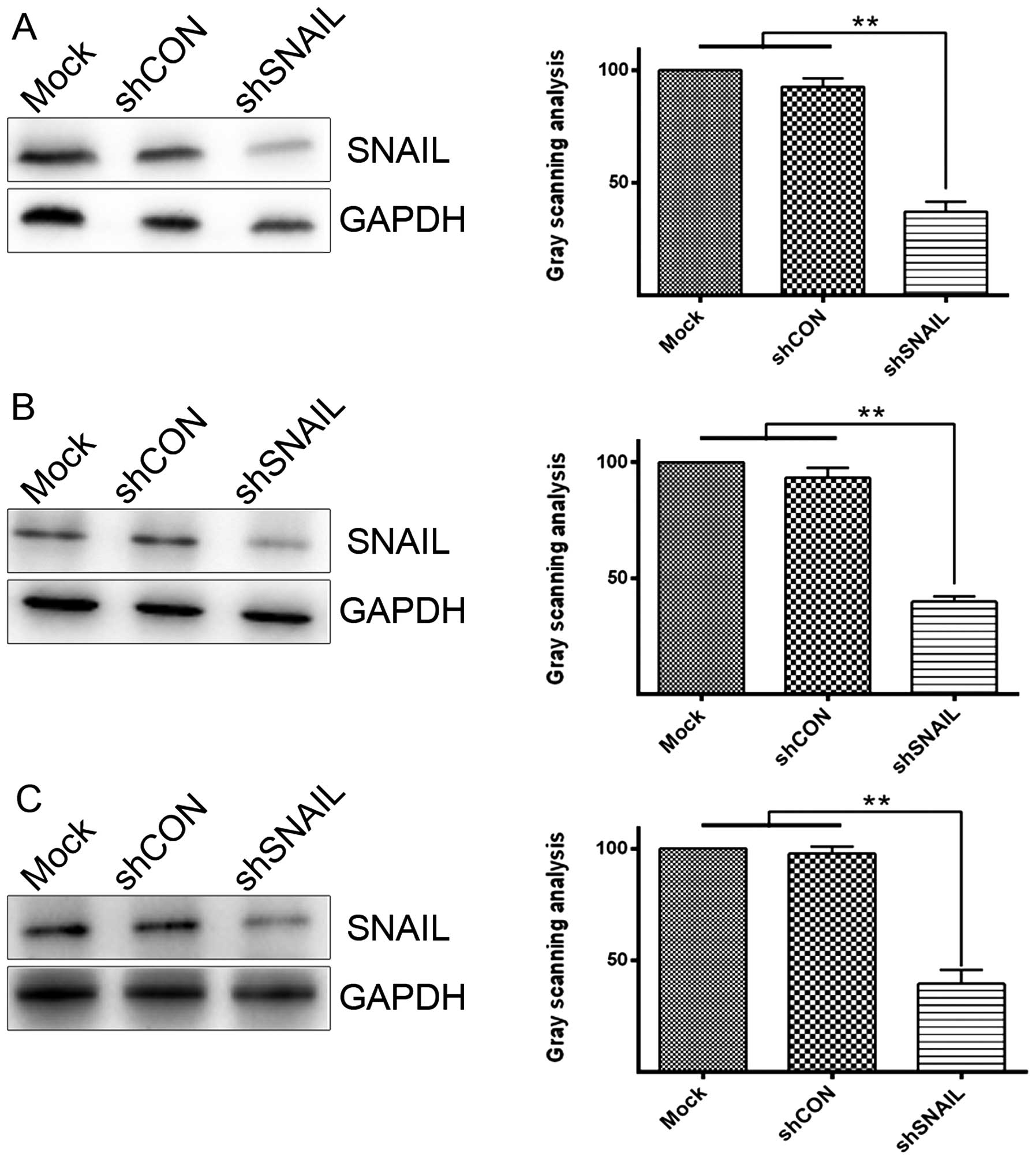
Expression of SNAIL in HCC cells
In our previous study (20), we detected the expression of SNAIL
in HepG2. Based on the results, LV-shSNAIL was found to suppress
the expression of SNAIL at the mRNA and protein level in HepG2
cells (20). The mRNA level was
measured by employing quantitative PCR, while western blot analysis
was used to determine the protein expression level. The results
verified that the level of SNAIL was obviously reduced following
LV-shSNAIL infection in other HCC cells (Fig. 1), which was consistent with the
anterior study (20). The findings
showed that the construction of LV-shSNAIL was successful in
silencing SNAIL expression in various types of HCC cells.
Cell viability and apoptosis analysis in
HCC cells
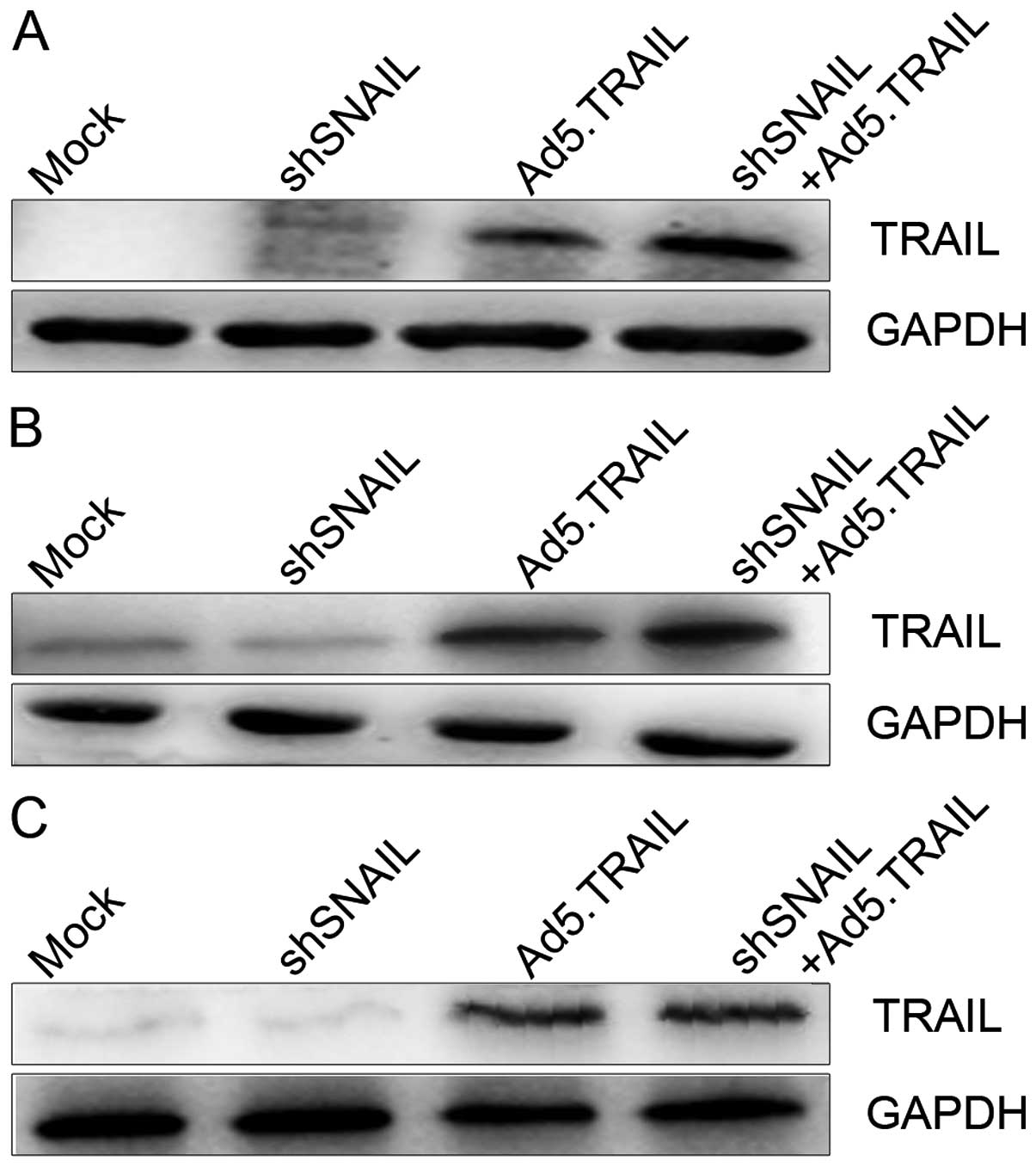
Recent findings have shown that, suppressing SNAIL
may inhibit invasion and metastasis in human HCC (26). Moreover, as described above,
silencing SNAIL reduced viability in HCC cells. It is well known
that the main function of TRAIL is the induction of tumor
apoptosis, as identified by findings from our laboratory (25,27).
Therefore, we first evaluated the status of TRAIL expression in
different groups of three types of HCC cells, and the level of
TRAIL was detected among the cells using western blot analysis.
Analysis of the results showed that TRAIL expression was markedly
increased following the addition of Ad5.TRAIL + LV-shSNAIL
(Fig. 2). These data showed that
TRAIL was effectively expressed following Ad5.TRAIL vector
infection of HCC cells, and that knockdown of SNAIL increased TRAIL
expression.
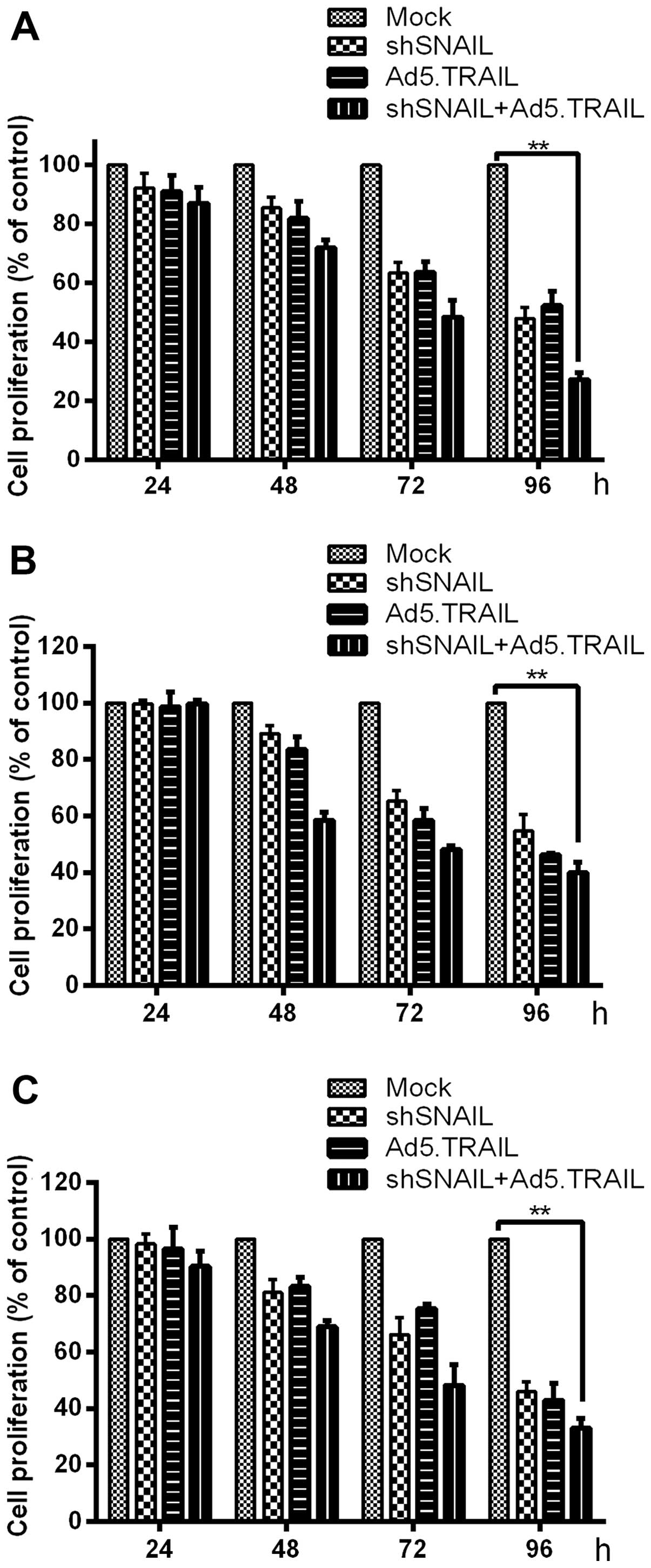
We also examined the effect of SNAIL and TRAIL on
HCC by assessing the state of cell proliferation. In order to study
the change in HCC cell viability for response to the SNAIL
silencing and TRAIL, respectively, we detected cell viability in
the four groups, from the point when reagents were added until 96
h, once every 24 h by MTT assay (Fig.
3). The extent of inducing apoptosis was also investigated. The
samples were stained with Hoechst 33258 for 48 h and then analyzed
by fluorescence microscopy (Fig.
4). The results revealed that silencing SNAIL or upregulated
TRAIL decreased proliferation and increased apoptosis in HCC
cells.
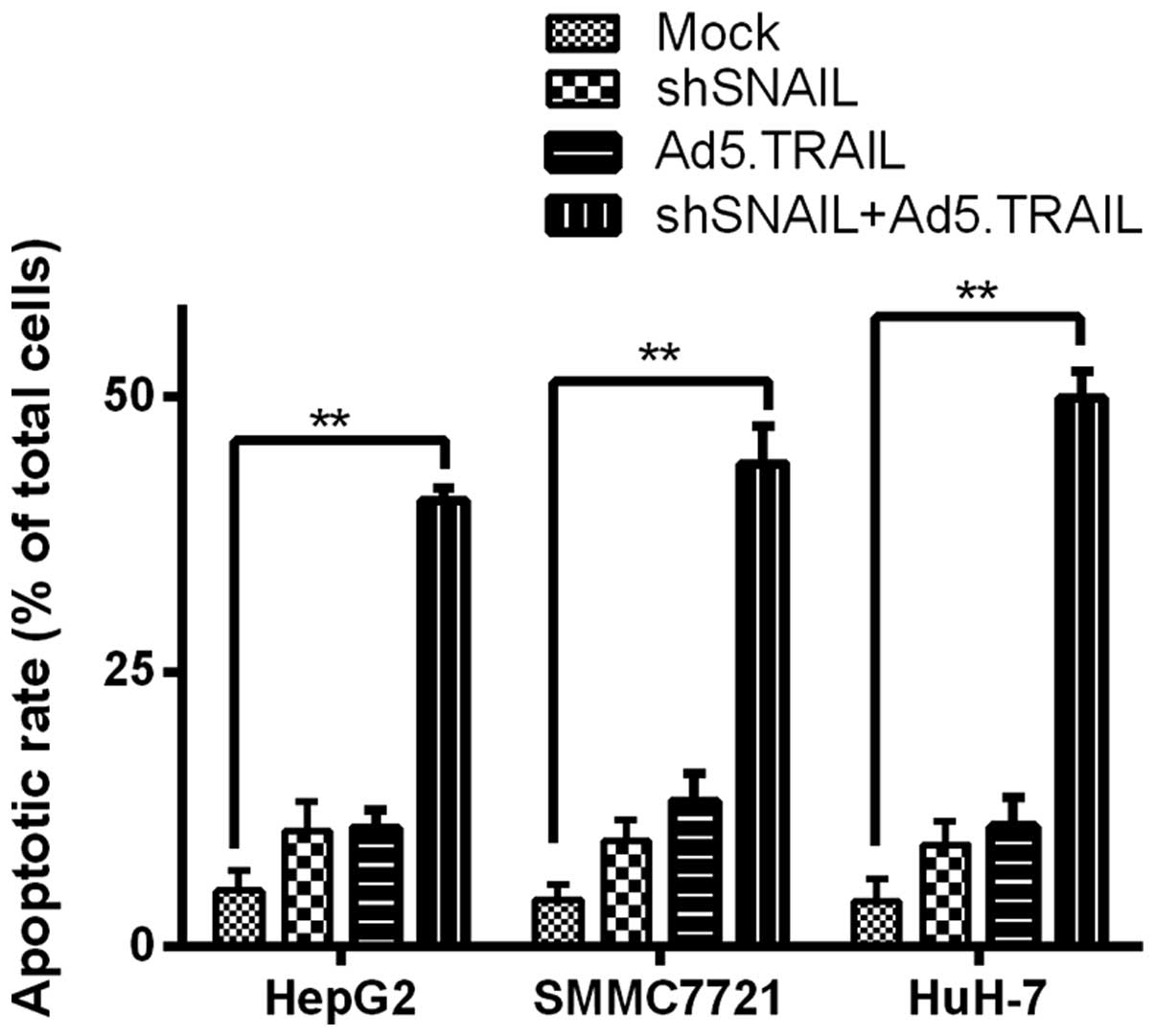
Inhibition of SNAIL enhanced
TRAIL-induced apoptosis
Since TRAIL-induced apoptosis was weakened in HCC
cells as compared to other TRAIL-sensitive cells, previous studies
focused on apoptosis induction by acting on TRAIL for HCC cells
(28). Therefore, we examined the
change of TRAIL-induced apoptosis in different groups. As shown in
Fig. 4, HCC cell apoptosis was
highest in the group infected with LV-shSNAIL and Ad5.TRAIL
compared to the other groups. This result suggested that silencing
SNAIL enhanced sensitivity that TRAIL induced HCC cell
apoptosis.
Knockdown of SNAIL increased
TRAIL-induced apoptosis by upregulating p53 expression
Previous authors reported that p53 sensitized
TRAIL-induced apoptosis in various types of cancer (29,30)
and blocking SNAIL activated p53, which regulated apoptosis
(31). To determine whether p53 was
associated with silencing of SNAIL to increase TRAIL-induced
apoptosis in HCC, we detected the expression level of p53 in
different groups in HuH-7 cells. The results revealed that p53
expression was highest in groups to which LV-shSNAIL was added than
those where LV-shSNAIL was not added (Fig. 5).
SNAIL silencing enhanced TRAIL-induced
apoptosis by affecting the NF-κB pathway
Resistance of HCC cells to TRAIL-induced apoptosis
has been shown to be associated with Bcl-xL, cIAP2 and survivin
factors, which are located in the downstream gene of the NF-κB
pathway (14,15,32).
Moreover, Hall et al suggested that blocking the activation
of Raf kinase inhibited NF-κB binding to the Mcl-1 promoter thereby
enhancing the sensitization of TRAIL-mediated apoptosis in cancer
cells (13). Among them, Raf-1
kinase, a member of the Raf kinase family, is an important
signaling molecule acting on the downstream kinases MEK and ERK,
which pathway converges on NF-κB activation (13,33).
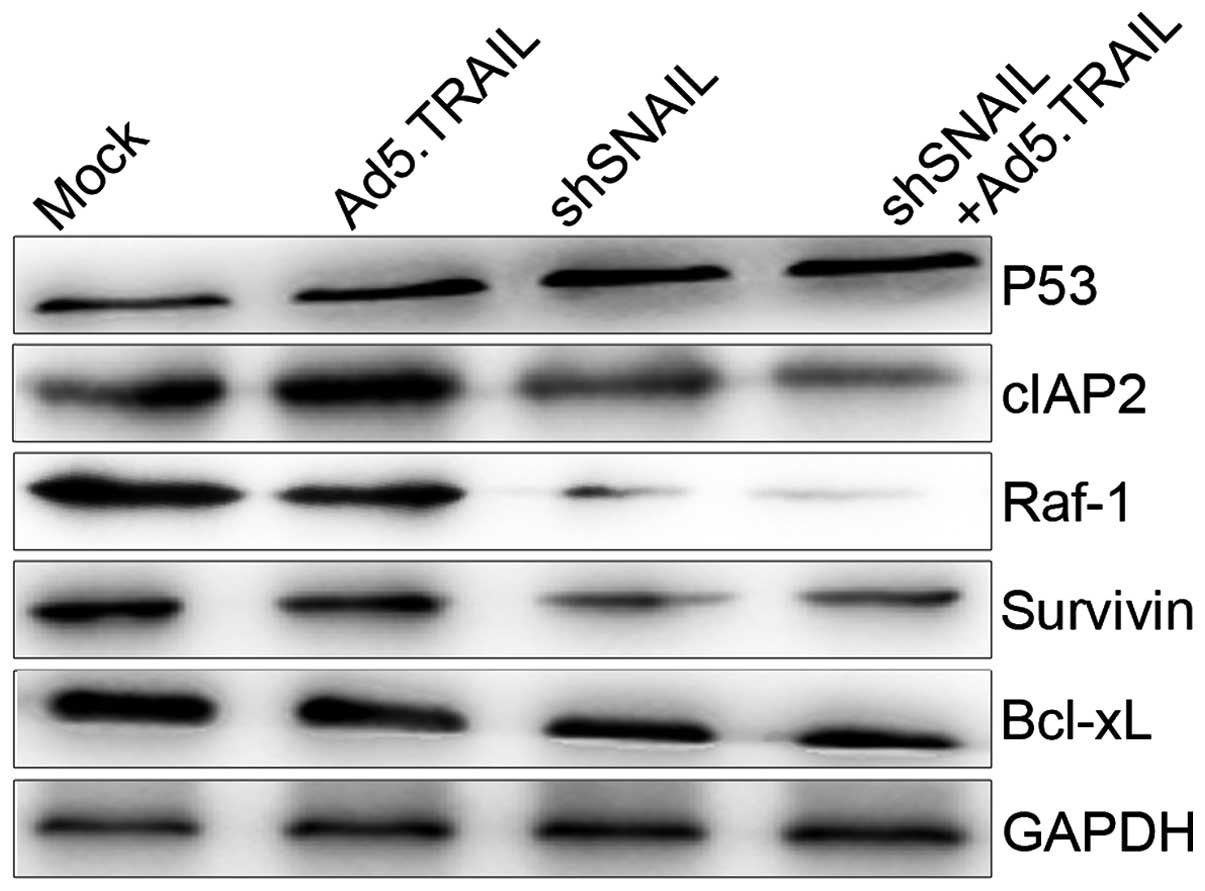
Accordingly, we investigated the expression levels of these
proteins in HCC cells to analyze the mechanism associated with the
enhancement of TRAIL-induced apoptosis by SNAIL silencing. The
results showed that Bcl-xL, cIAP2, survivin and Raf-1 expression
were reduced following the addition of LV-shSNAIL than without its
addition to HuH-7 cells, and particularly low in LV-shSNAIL +
Ad5.TRAIL (Fig. 5).
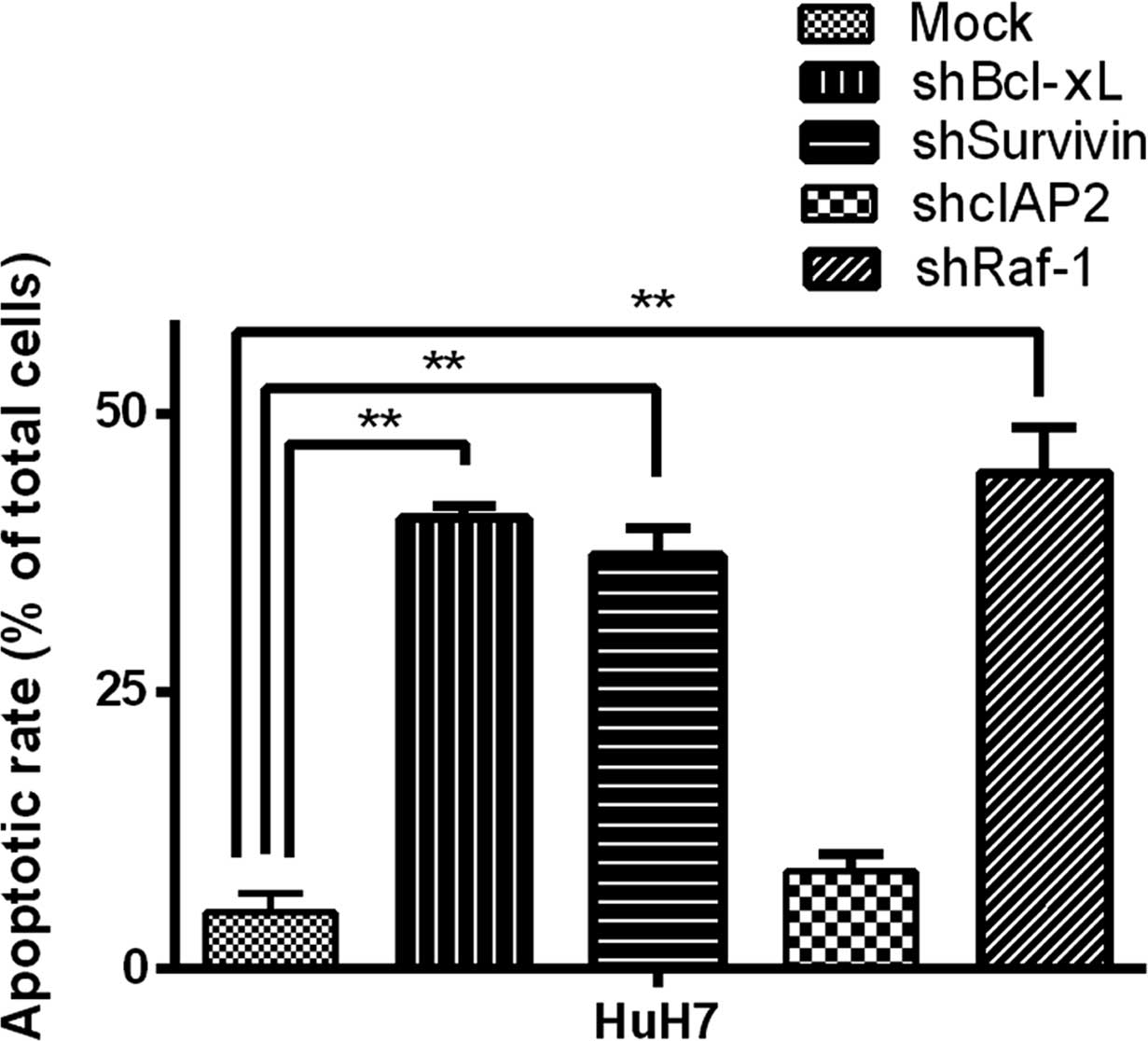
In order to determine whether Bcl-xL, cIAP2,
survivin and Raf-1 participated in the suppression of SNAIL to
mediate HCC cell sensitivity to TRAIL-induced apoptosis, we used
lentiviral vectors carrying shRNA to infect HuH-7 for the knockdown
of Bcl-xL, cIAP2, survivin and Raf-1. The effect of TRAIL-induced
apoptosis of HuH-7 cells was analyzed by fluorescence microscopy
following the knockdown of each gene. Infection of LV-shBcl-xL,
LV-shSurvivin or LV-shRaf-1 HuH-7 cells showed obvious sensitivity
to TRAIL-induced apoptosis, whereas the sensitivity of shcIAP2 was
not affected (Fig. 6).
In conclusion, Bcl-xL, survivin and Raf-1
downregulation was conducive to enhancement of HCC cell sensitivity
to TRAIL-induced apoptosis by silencing SNAIL. In other words,
silencing of SNAIL enhanced TRAIL-induced apoptosis by affecting
the NF-κB pathway.
Discussion
In the present study, shSNAIL carrying lentiviral
and adenoviral vectors harboring TRAIL induced SNAIL silencing and
enhanced TRAIL expression. The results show that HCC cell apoptosis
was increased whether via silencing SNAIL or enhancing the level of
TRAIL, as confirmed by the cell viability results. The results also
show that the sensitivity of TRAIL-induced apoptosis was increased
when SNAIL was silenced. To examine the mechanism involved, we
found that p53 performed a crucial role in the sensitization of HCC
cells to TRAIL by downregulating SNAIL. Other mechanisms identified
showed that silencing SNAIL enhanced TRAIL-induced apoptosis by
affecting the NF-κB pathway. Therefore, enhancing the sensitization
of HCC cells to TRAIL is essential for the promotion of HCC cell
apoptosis. Moreover, SNAIL is a potential target that may overcome
resistance of HCC cells to TRAIL-induced apoptosis, rendering it an
improved therapeutic strategy for HCC.
As previously mentioned, most HCC cells were
resistant to apoptosis in many stimuli, which included TRAIL.
However, findings of recent studies (3,12,13,30)
have shown that many targeted drugs alter their characteristics by
acting on special targets. In the present study, we found that
silencing SNAIL can increase the expression of p53, which alters
the sensitization of HCC cells to TRAIL. On the other hand, it has
been reported that p53 upregulated TRAIL death receptors (34,35).
Our results have shown that silencing SNAIL upregulated p53.
However, whether TRAIL death receptors are correlated with p53
remains to be determined. In addition, another study has shown that
adenoviral-mediated transfer of p53 gene enhanced
TRAIL-induced apoptosis in human HCC cells, not because of the
induction of TRAIL death receptors, but due to the downregulation
of cFLIP or XIAP (36), which
resisted sensitization of some cell types to TRAIL-induced
apoptosis (32,37). Thus p53 is an important gene
that enhances the sensitization of HCC cells to TRAIL by
downregulating cFLIP or XIAP under certain stimulation.
Irrespective of the mechanism involved, p53 as a target is an
important breakthrough point of HCC treatment.
Bcl-xL as an anti-apoptosis-associated protein
member of the Bcl-2 family may play a very vital role in regulating
the apoptosis of HCC (38).
Overexpression of Bcl-xL contributes to TRAIL resistance in cancer,
including HCC (14,38–40).
Given that Bcl-xL may be a bridge between SNAIL and TRAIL-mediated
apoptosis in HCC, we further studied the relationship among them.
The result showed that the expression of Bcl-xL was reduced by
silencing SNAIL in HCC, and according to our experimental data,
suppression of Bcl-xL by SNAIL silencing was able to increase the
sensitivity of TRAIL-induced HCC cell apoptosis.
Similarly, previous studies have shown that the
apoptotic inhibitors of cIAP2 and survivin are important factors in
determining the apoptosis of HCC cells (3,14).
Thus, we investigated the expression of the two proteins. The
results showed that cIAP2 and survivin were downregulated after
silencing SNAIL. Additionally, we found that a decrease of survivin
expression not only induced HCC cell apoptosis augment, but also
enhanced the sensitivity of TRAIL-mediated apoptosis in HCC cells
following the knockdown of SNAIL. However, cIAP2 did not affect
TRAIL-induced apoptosis. Nevertheless, the exact mechanism on how
these proteins are regulated by SNAIL silencing remains to be
clarified.
A previous study demonstrated the crucial role of
SNAIL in the resistance of tumor cells to TRAIL (41). As a downstream target of TRAIL, many
factors acted on this role of SNAIL. Furthermore, our results show
that, SNAIL is important in mediating these factors, exerting an
indirect effect. SNAIL has been shown to suppress Raf-kinase
inhibitor protein (RKIP) transcription and expression, which was
consistent with our observation that Raf-1 protein was
downregulated after SNAIL silencing, and previous findings have
shown that RKIP inhibited NF-κB activity (42,43).
Another study showed that NF-κB induces the anti-apoptotic
regulator Mcl-1, which was able to cause TRAIL-resistance for
cancer (13). Therefore, the
potential mechanism was that silencing SNAIL could indirectly
decrease Mcl-1 to mediate the sensitization of TRAIL-induced
apoptosis in HCC cells by upregulating RKIP and downregulating
NF-κB. Thus, silencing SNAIL down-regulated Raf protein to
influence NF-κB activity, thereby sensitizing HCC cells to TRAIL,
which has been confirmed in our study. Nevertheless, the mechanisms
involved remain to be elucidated.
In conclusion, SNAIL is a vital mediator of HCC cell
sensitivity to TRAIL-induced apoptosis. Inhibition of SNAIL through
shRNA carried by lentivirus is an important method that reversed
HCC cell resistance to TRAIL by affecting the NF-κB pathway.
Therefore, SNAIL can be a significant gene for treating HCC and
identification of a new target to change the sensitization of
TRAIL-induced apoptosis in HCC is an important therapeutic
strategy.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundations of China (nos. 81372632 and 81402579)
and the Postdoctorate Innovation Foundation of Shandong Province of
China (no. 201303063).
References
|
1
|
Marquardt JU and Thorgeirsson SS:
SnapShot: hepatocellular carcinoma. Cancer Cell. 25:5502014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamanaka Y, Shiraki K, Inoue T, Miyashita
K, Fuke H, Yamaguchi Y, Yamamoto N, Ito K, Sugimoto K and Nakano T:
COX-2 inhibitors sensitize human hepatocellular carcinoma cells to
TRAIL-induced apoptosis. Int J Mol Med. 18:41–47. 2006.PubMed/NCBI
|
|
4
|
Chun E and Lee KY: Bcl-2 and Bcl-xL are
important for the induction of paclitaxel resistance in human
hepatocellular carcinoma cells. Biochem Biophys Res Commun.
315:771–779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ogunwobi OO and Liu C: Hepatocyte growth
factor upregulation promotes carcinogenesis and
epithelial-mesenchymal transition in hepatocellular carcinoma via
Akt and COX-2 pathways. Clin Exp Metastasis. 28:721–731. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang Y, Zhu J, Gou H, Cao D, Jiang M and
Hou M: Clinical significance of Cox-2, Survivin and Bcl-2
expression in hepatocellular carcinoma (HCC). Med Oncol.
28:796–803. 2011. View Article : Google Scholar
|
|
7
|
Deng Q, Zhang Z, Feng X, et al:
TRAIL-secreting mesenchymal stem cells promote apoptosis in
heat-shock-treated liver cancer cells and inhibit tumor growth in
nude mice. Gene Ther. 21:317–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashkenazi A and Dixit VM: Apoptosis
control by death and decoy receptors. Curr Opin Cell Biol.
11:255–260. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Benedict CA and Ware CF: TRAIL: not just
for tumors anymore? J Exp Med. 209:1903–1906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garimella SV, Gehlhaus K, Dine JL, Pitt
JJ, Grandin M, Chakka S, Nau MM, Caplen NJ and Lipkowitz S:
Identification of novel molecular regulators of tumor necrosis
factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis
in breast cancer cells by RNAi screening. Breast Cancer Res.
16:R412014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamanaka T, Shiraki K, Sugimoto K, Ito T,
Fujikawa K, Ito M, Takase K, Moriyama M, Nakano T and Suzuki A:
Chemotherapeutic agents augment TRAIL-induced apoptosis in human
hepatocellular carcinoma cell lines. Hepatology. 32:482–490. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ganten TM, Koschny R, Haas TL, Sykora J,
Li-Weber M, Herzer K and Walczak H: Proteasome inhibition
sensitizes hepatocellular carcinoma cells, but not human
hepatocytes, to TRAIL. Hepatology. 42:588–597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hall MA and Cleveland JL: Clearing the
TRAIL for cancer therapy. Cancer Cell. 12:4–6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Omar HA, Arafa el-SA, Maghrabi IA and Weng
JR: Sensitization of hepatocellular carcinoma cells to Apo2L/TRAIL
by a novel Akt/NF-κB signalling inhibitor. Basic Clin Pharmacol
Toxicol. 114:464–471. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Refaat A, Abd-Rabou A and Reda A: TRAIL
combinations: The new ‘trail’ for cancer therapy (Review). Oncol
Lett. 7:1327–1332. 2014.PubMed/NCBI
|
|
16
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: an alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pioli PD and Weis JH: Snail transcription
factors in hematopoietic cell development: a model of functional
redundancy. Exp Hematol. 42:425–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kudo-Saito C, Shirako H, Takeuchi T and
Kawakami Y: Cancer metastasis is accelerated through
immunosuppression during Snail-induced EMT of cancer cells. Cancer
Cell. 15:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyoshi A, Kitajima Y, Kido S, Shimonishi
T, Matsuyama S, Kitahara K and Miyazaki K: Snail accelerates cancer
invasion by upregulating MMP expression and is associated with poor
prognosis of hepatocellular carcinoma. Br J Cancer. 92:252–258.
2005.PubMed/NCBI
|
|
20
|
Liu J, Jiang G, Liu S, Liu Z, Pan H, Yao R
and Liang J: Lentivirus-delivered short hairpin RNA targeting SNAIL
inhibits HepG2 cell growth. Oncol Rep. 30:1483–1487.
2013.PubMed/NCBI
|
|
21
|
Fellmann C and Lowe SW: Stable RNA
interference rules for silencing. Nat Cell Biol. 16:10–18. 2014.
View Article : Google Scholar :
|
|
22
|
Takakura Y: Towards therapeutic
application of RNA-mediated gene regulation. Preface Adv Drug Deliv
Rev. 61:6672009. View Article : Google Scholar
|
|
23
|
Djuranovic S, Nahvi A and Green R: A
parsimonious model for gene regulation by miRNAs. Science.
331:550–553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jinek M and Doudna JA: A three-dimensional
view of the molecular machinery of RNA interference. Nature.
457:405–412. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Wang J, Wang H, Liu S, Liang Y, Lv
Z, Zhou Q and Ding W: Combination of Ad-sTRAIL with the
chemotherapeutic drug cisplatin synergistically enhances their
pro-apoptotic ability in human breast cancer cells. Oncol Rep.
30:1913–1919. 2013.PubMed/NCBI
|
|
26
|
Chen D, Zheng X, Jiao X, Gao Y, Zhang K
and Liang J: Transcriptional repressor snail and metastasis in
hepatocellular carcinoma. Hepatogastroenterology. 59:1359–1365.
2012.
|
|
27
|
Smyth MJ, Takeda K, Hayakawa Y, Peschon
JJ, van den Brink MR and Yagita H: Nature’s TRAIL - on a path to
cancer immunotherapy. Immunity. 18:1–6. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Abou El Naga RN, Azab SS, El-Demerdash E,
Shaarawy S, El-Merzabani M and Ammar el-SM: Sensitization of
TRAIL-induced apoptosis in human hepatocellular carcinoma HepG2
cells by phytochemicals. Life Sci. 92:555–561. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu X, Qiu F, Liu Z, Lan Y, Wang K, Zhou
PK and Wang Y: Urokinase-type plasminogen activator receptor
regulates apoptotic sensitivity of colon cancer HCT116 cell line to
TRAIL via JNK-p53 pathway. Apoptosis. 19:1532–1544. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao J, Lu Y and Shen HM: Targeting p53 as
a therapeutic strategy in sensitizing TRAIL-induced apoptosis in
cancer cells. Cancer Lett. 314:8–23. 2012. View Article : Google Scholar
|
|
31
|
Lee SH and Park BJ: p53 activation by
blocking Snail: a novel pharmacological strategy for cancer. Curr
Pharm Des. 17:610–617. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Finlay D, Vamos M, Gonzalez-Lopez M, et
al: Small-molecule IAP antagonists sensitize cancer cells to
TRAIL-induced apoptosis: roles of XIAP and cIAPs. Mol Cancer Ther.
13:5–15. 2014. View Article : Google Scholar :
|
|
33
|
Dougherty MK, Müller J, Ritt DA, Zhou M,
Zhou XZ, Copelan TD, Conrads TP, Veenstra TD, Lu KP and Morrison
DK: Regulation of Raf-1 by direct feedback phosphorylation.
17:215–224. 2005.
|
|
34
|
Akram KM, Lomas NJ, Forsyth NR and Spiteri
MA: Alveolar epithelial cells in idiopathic pulmonary fibrosis
display upregulation of TRAIL, DR4 and DR5 expression with
simultaneous preferential over-expression of pro-apoptotic marker
p53. Int J Clin Exp Pathol. 7:552–564. 2014.PubMed/NCBI
|
|
35
|
Yeh CH, Yang YY, Huang YF, Chow KC and
Chen MF: Induction of apoptosis in human Hep3B hepatoma cells by
norcantharidin through a p53 independent pathway via TRAIL/DR5
signal transduction. Chin J Integr Med. 18:676–682. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Inoue H, Shiraki K, Murata K, et al:
Adenoviral-mediated transfer of p53 gene enhances TRAIL-induced
apoptosis in human hepatocellular carcinoma cells. Int J Mol Med.
14:271–275. 2004.PubMed/NCBI
|
|
37
|
Cantarella G, Di Benedetto G, Ribatti D,
Saccani-Jotti G and Bernardini R: Involvement of caspase 8 and
c-FLIPL in the proangiogenic effects of the tumour necrosis
factor-related apoptosis-inducing ligand (TRAIL). FEBS J.
281:1505–1513. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo XZ, Shao XD, Liu MP, Xu JH, Ren LN,
Zhao JJ, Li HY and Wang D: Effect of bax, bcl-2 and bcl-xL on
regulating apoptosis in tissues of normal liver and hepatocellular
carcinoma. World J Gastroenterol. 8:1059–1062. 2002.PubMed/NCBI
|
|
39
|
Bansal H, Seifert T, Bachier C, Rao M,
Tomlinson G, Iyer SP and Bansal S: The transcription factor Wilms
tumor 1 confers resistance in myeloid leukemia cells against the
proapoptotic therapeutic agent TRAIL (tumor necrosis factor
alpha-related apoptosis-inducing ligand) by regulating the
antiapoptotic protein Bcl-xL. J Biol Chem. 287:32875–32880. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Koehler BC, Urbanik T, Vick B, Boger RJ,
Heeger S, Galle PR, Schuchmann M and Schulze-Bergkamen H:
TRAIL-induced apoptosis of hepatocellular carcinoma cells is
augmented by targeted therapies. World J Gastroenterol.
15:5924–5935. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaler P, Galea V, Augenlicht L and
Klampfer L: Tumor associated macrophages protect colon cancer cells
from TRAIL-induced apoptosis through IL-1beta-dependent
stabilization of Snail in tumor cells. PLoS One. 5:e117002010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Baritaki S and Bonavida B: Viral infection
and cancer: the NF-kappaB/Snail/RKIP loop regulates target cell
sensitivity to apoptosis by cytotoxic lymphocytes. Crit Rev
Immunol. 30:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu K and Bonavida B: The activated
NF-kappaB-Snail-RKIP circuitry in cancer regulates both the
metastatic cascade and resistance to apoptosis by cytotoxic drugs.
Crit Rev Immunol. 29:241–254. 2009. View Article : Google Scholar : PubMed/NCBI
|