Introduction
Cancer stem cells (CSCs) exist within tumors and are
a unique subset of cells with the potential to self-renew,
proliferate indefinitely, and differentiate into tumor cells
(1,2). Present cancer therapies are known to
leave behind some CSCs, explaining why tumor eradication is
difficult to achieve (3–5). While the development of CSC-specific
drugs may bring new hope to cancer therapy, there are few
established models that allow the isolation and the study of
CSCs.
Previous findings have shown that aldehyde
dehydrogenase 1 (ALDH1) can be used as a CSC marker and is
expressed in many stem cell types, including breast, lung,
neuronal, and hematopoietic stem cells (HSCs) (6,7). ALDH1
is a member of a family of acetaldehyde dehydrogenase enzymes that
oxidize acetaldehyde to acetic acid. In HSCs, ALDH1 can control
retinol metabolism, and certain classes of retinoic acid may lead
to the differentiation of hematopoietic progenitor cells (8). By contrast, immature HSCs are very
rich in retinol, which enhances self-renewal (9). The above-mentioned results suggest
that ALDH1 is a necessary factor for the HSC environment. A
fluorescent ALDH substrate that passes through the cell membrane by
free diffusion (Aldefluor® BAAA-ALDH) can be used to
measure ALDH enzyme activity by quantitative flow cytometry
(10–12).
Malignant fibrous histiocytoma (MFH) was identified
and designated as such in the 1960s. However, the prognostic
factors that are predictive of MFH outcome have not been well
described (13). The majority of
MFH tumors are defined as pleomorphic subtypes, and a few myxoid
MFH cell lines have been established (14–16).
Kawashima et al established the NMFH-1 cell line, which may
prove a useful tool for studying CSCs (17).
In the present study, we isolated and characterized
the population of NMFH-1 cells that have ALDH enzymatic activity, a
trait characteristic of CSCs. The aim was to develop a model for
the study of CSCs, which may ultimately provide insights into the
clinical treatment of MFH. At present, it remains unknown whether
the population of ALDH+ cells may be isolated from the
human NMFH-1 cell line. Therefore, we defined the population of
NMFH-1 cells with high ALDH enzymatic activity on CSC phenotypes.
The findings may impact on the development of more effective MFH
therapies.
Materials and methods
Cell lines and culture
The human NMFH-1 cell line used in the present study
was obtained from the Niigata University Graduate School of Medical
and Dental Sciences (17) and
maintained at 37°C in a 5% CO2 atmosphere in RPMI-1640
supplemented with 10% fetal bovine serum (FBS).
Aldefluor® (Stem Cell Technologies, Durham, NC, USA) was
used to isolate the cell populations with high levels of the ALDH
enzymatic activity. The cells were labeled with
Aldefluor® reagent by collecting cells using 0.25%
trypsin according to the manufacturer’s instructions, and
single-cell suspension was prepared. The cells were washed with PBS
and isolated by centrifugation prior to adding the product reagent
according to the manufacturer’s instructions. The ALDH+
and ALDH− cells were then analyzed or isolated using a
FACSAria.
Sphere formation
A single-cell suspension of 1×105
ALDH+ or ALDH− cells in serum-free RPMI-1640
medium was seeded in 6-well plates (Corning Inc., Corning, New
York, USA). Each well contained 20 μg/l of EGF and bFGF, and
wells were supplemented with 2 μg/l of EGF and bFGF every 24
h until spherical cell formations appeared. Following sphere
formation, the cells were collected in a common culture bottle with
complete media to observe whether the cells could be adhered to the
wall. Other spheres were disrupted into a signal-cell solution to
assess whether a second sphere could form.
Tumor implantation in nude BALB/c
mice
Cells were collected and suspended and the
concentration was adjusted with culture medium. The cells were
injected into the left side of the front leg of BALB/c nude mice (6
weeks; weight, 18–22 g) obtained from the Animal Research Center,
Harbin Medical University, China. The mice were separated into 10
groups of 5 animals and received varying concentrations of the
ALDH+ or ALDH− cells. After 6 weeks, the mice
were sacrificed and any tumors were removed for histopathological
and immunohistochemical analysis. Tumor samples were also digested
using collagenase II (Sigma-Aldrich, St. Louis, MO, USA) and
re-injected into mice to generate second-round tumors. Data were
collected from three independent experiments.
Histopathological and immunohistochemical
analysis of xenografts
The ALDH+ and ALDH− tumors
were placed in flasks with 10% formalin, and then embedded in
paraffin. Sections were cut and stained with hematoxylin and eosin
(H&E) using a standard protocol to assess tumor type. An ALDH1
antibody (Cell Signaling Technology, Danvers, MA, USA) was used to
determine the ALDH1 expression levels in the tumor by
immunohistochemistry.
Chemoresistance of cell monolayers and
spheres
Cells were transferred to 96-well plates at a
density of 2×103 cells/well in RPMI-1640 culture medium
with 10% FBS. Increasing concentrations of the drugs doxorubicin
(DXR) or cisplatin (CDDP) (Sigma-Aldrich) were added in triplicate
and incubated for 40 h (1, 5 and 10 μM). CCK-8 (10
μl) was placed in each well. The OD value was measured at
450 nm. Cell viability was measured using the Cell Titer 96 AQueous
One Solution Cell Proliferation Assay kit (Promega), according to
the manufacturer’s instructions. In addition, we compared drug
resistance between the ALDH+ spheroid and adherent
cells.
Quantitative PCR (qPCR)
Total RNA was extracted from both the
ALDH+ and ALDH− cells. We quantified the mRNA
transcription levels of highly expressed genes in stem cells, i.e.,
c-Myc, Bmi-1, Sox2, Nanog,
Oct3/4, and STAT3. We also assessed the expression
levels of the drug-resistant genes, ABCG2 and ALDH1.
β-actin was used as an internal control. The primer sequences used
for amplification are listed in Table
I.
 | Table IPCR primers used in this study. |
Table I
PCR primers used in this study.
| Target gene | Primer
sequences | Size (bp) |
|---|
| c-Myc | F:
5′-TCCCTCCACTCGGAAGGAC-3′
R: 5′-CTGGTGCATTTTCGGTTGTTG-3′ | 96 |
| Nanog | F:
5′-TTTGTGGGCCTGAAGAAAACT-3′
R: 5′-AGGGCTGTCCTGAATAAGCAG-3′ | 116 |
| Sox-2 | F:
5′-GCCGAGTGGAAACTTTTGTCG-3′
R: 5′-GGCAGCGTGTACTTATCCTTCT-3′ | 155 |
| Bmi-1 | F:
5′-CGTGTATTGTTCGTTACCTGGA-3′
R: 5′-TTCAGTAGTGGTCTGGTCTTGT-3′ | 82 |
| ABCG2 | F:
5′-ACGAACGGATTAACAGGGTCA-3′
R: 5′-CTCCAGACACACCACGGAT-3′ | 93 |
| STAT3 | F:
5′-CAGCAGCTTGACACACGGTA-3′
R: 5′-AAACACCAAAGTGGCATGTGA-3′ | 150 |
| ALDH1 | F:
5′-GCACGCCAGACTTACCTGTC-3′
R: 5′-CCTCCTCAGTTGCAGGATTAAAG-3′ | 129 |
| Oct3/4 | F:
5′-GTGTTCAGCCAAAAGACCATCT-3′
R: 5′-GGCCTGCATGAGGGTTTCT-3′ | 156 |
| β-actin | F:
5′-CATGTACGTTGCTATCCAGGC-3′
R: 5′-CTCCTTAATGTCACGCACGAT-3′ | 250 |
Western blotting
Western blotting was performed as described
previously, with some modifications (18). Briefly, the cells were washed twice
with cold PBS and lysed in buffer containing 50 mM Tris-HCl (pH
7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1% Na
deoxycholate, 1 mM Na vanadate and protease inhibitors (5 mg/ml
pepstatin, 1 mM PMSF, 10 mg/ml leupeptin and 1 mM NaF;
Sigma-Aldrich) for 1 h on ice. After centrifugation at 13,000 × g
for 10 min at 4°C, the supernatant protein concentrations were
measured using the BCA Protein Assay kit (Pierce Biotechnology,
Inc., Rockford, IL, USA). Membranes were blocked with non-fat milk
for 1 h at room temperature and incubated overnight at 4°C with the
corresponding antibodies in Tris-buffered saline (TBS) with 5% BSA
(Sigma-Aldrich) and 0.1% Tween-20 (Bio-Rad Laboratories, Hercules,
CA, USA). After washing three times in TBS with 0.1% Tween-20, the
blots were incubated with horseradish peroxidase-conjugated
secondary antibody (Cell Signaling Technology, Danvers, MA, USA;
Jackson ImmunoResearch Laboratories, West Grove, PA, USA).
Immunoreactive bands were detected with the ECL Plus SuperSignal
West Pico Chemiluminescent Substrate (Thermo Scientific, Waltham,
MA, USA) for 60 sec. Protein levels were normalized with respect to
the band density of glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) as an internal control. The primary antibodies used were:
anti-ABCG2 (1:1,000), anti-Bmi-1 (1:1,000), anti-c-Myc (1:1,000),
anti-Nanog (1:1,000), anti-Oct3/4 (1:1,000), anti-Sox2 (1:1,000),
anti-STAT3 (1:1,000), and anti-ALDH1 (1:1,000) (all from Cell
Signaling Technology), and anti-GAPDH (1:5,000) (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA).
Statistical analysis
The statistical software SPSS 20.0 (SPSS Inc.,
Chicago, IL, USA) was used for data processing and analyzing. Data
are presented as the means ± SD, and a comparison was carried
between experimental groups of qPCR analysis using one-way ANOVA.
The results of the chemosensitivity assay were calculated and
compared using one-way ANOVA. P<0.05 was considered
statistically significant.
Results
Separation and differentiation of the
ALDH+ and ALDH− populations from the human
NMFH-1 cell line
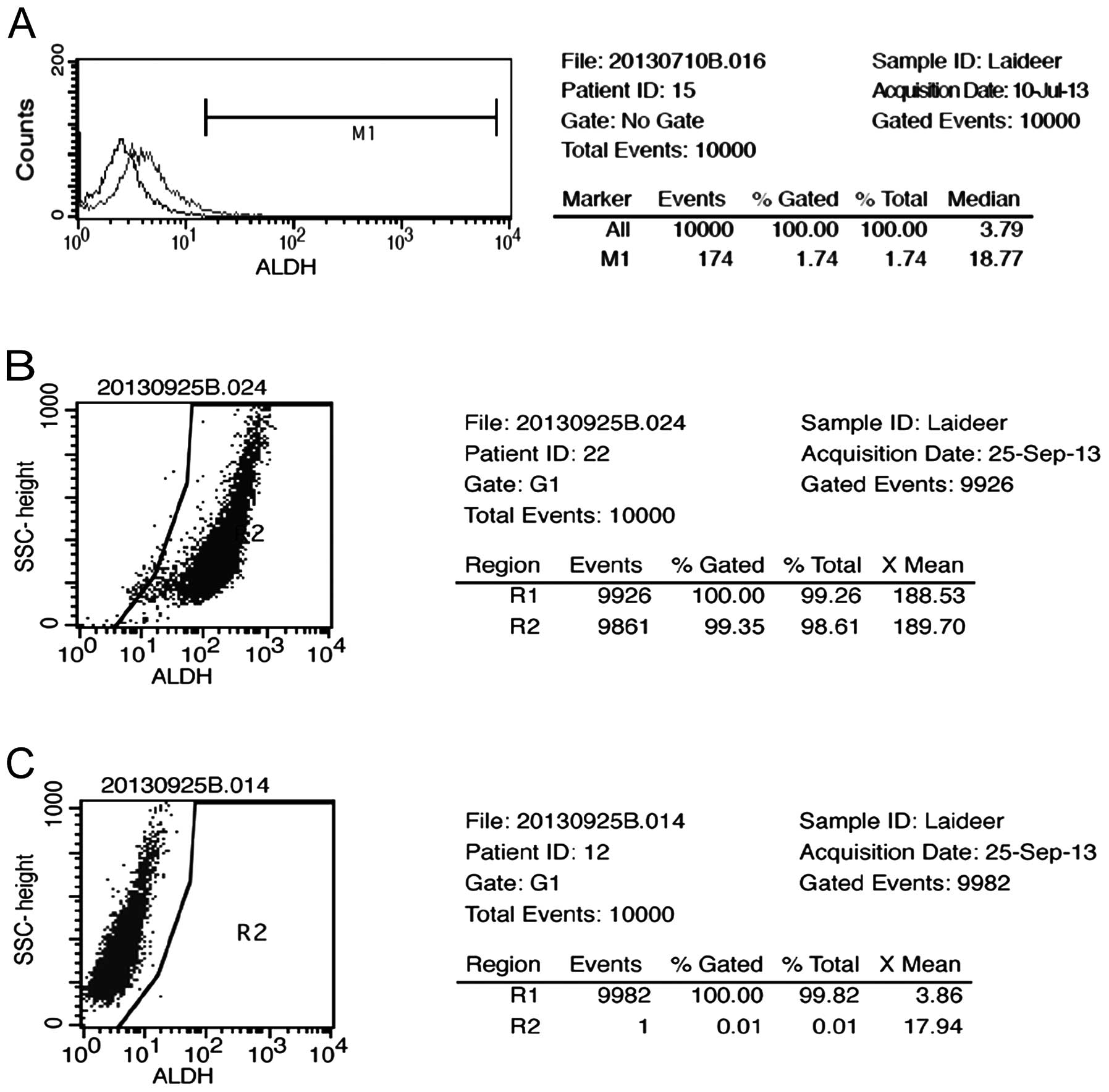
We assessed the presence and size of the NMFH-1 cell
population with the ALDH enzymatic activity by Aldefluor assay. A
small (1.74%) but detectable cell population was identified
(Fig. 1A). The ALDH+ and
ALDH− cells were isolated by FACSAria-based cell
sorting, and the content of each population was verified after
separation. The ALDH+ population reached 98.61% purity
(Fig. 1B), and the ALDH−
population reached 99.82% purity (Fig.
1C).
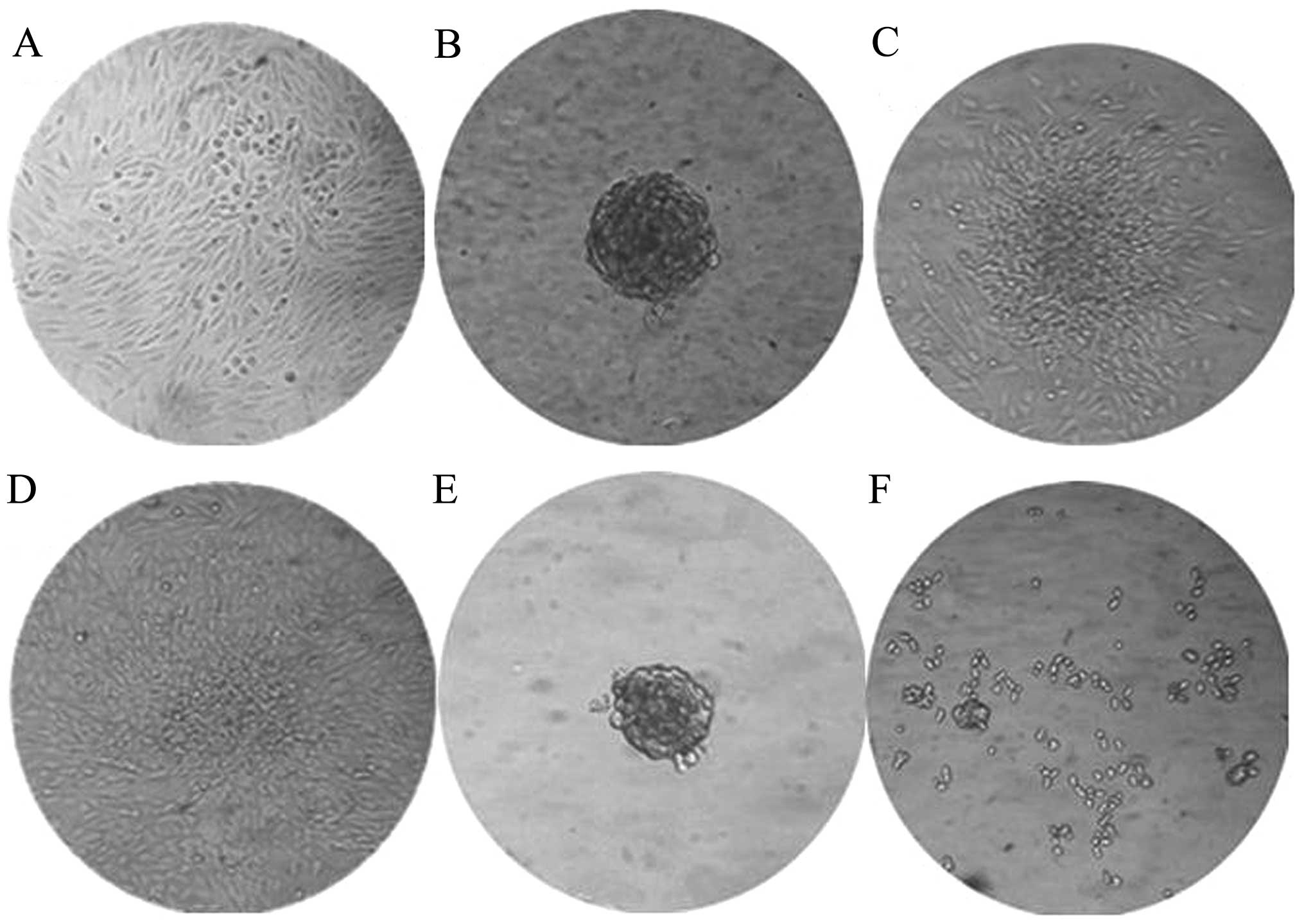
ALDH+ cells exhibit enhanced
sphere formation
The ALDH+ cells formed many spheroids
when cultured in suspension with serum-free medium. This method was
initially developed to select neural stem cells, but has been
adapted as a general tumor-initiating cell selection method. The
cells grew normally under these conditions (Fig. 2A), but growth in
EGF/bFGF-supplemented medium has been shown to allow spherical
clones to develop from normal cells and CSCs of epithelial origin.
The ALDH+ cells formed a small sphere on the third day,
which became clearly visible by the fifth day (Fig. 2B). In comparison, 10 days were
required for the ALDH− cells to develop spheres that
were smaller than those formed by the ALDH+ populations
(Fig. 2F). The ALDH+
spheres could be transferred into complete media containing serum
and the cells were able to adhere for continued growth (Fig. 2C and D).
ALDH+ cells exhibit enhanced
tumor development in BALB/c nude mice
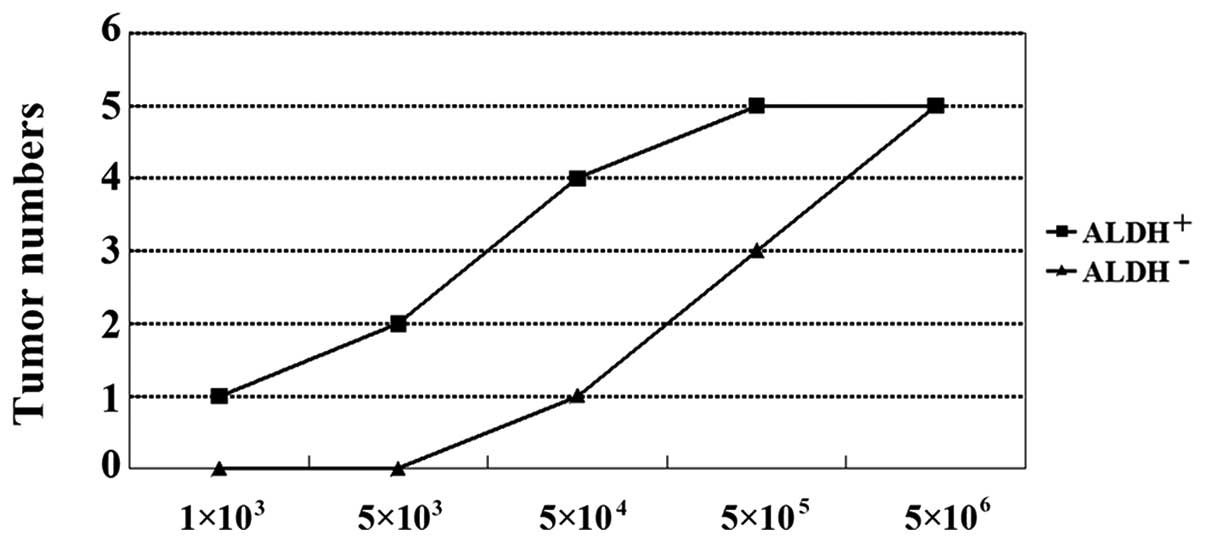
The implantation of cancer cells into nude mice can
lead to tumor development. In order to evaluate the tumor
development potential of the ALDH+ and ALDH−
cells, we injected them into BALB/c nude mice to generate
xenografts. Six weeks following the injection, the mice were
euthanized and assayed for tumor development (Table II). As few as 1×103
ALDH+-injected cells were required for tumor growth. By
contrast, 5×104 ALDH− cells were required to
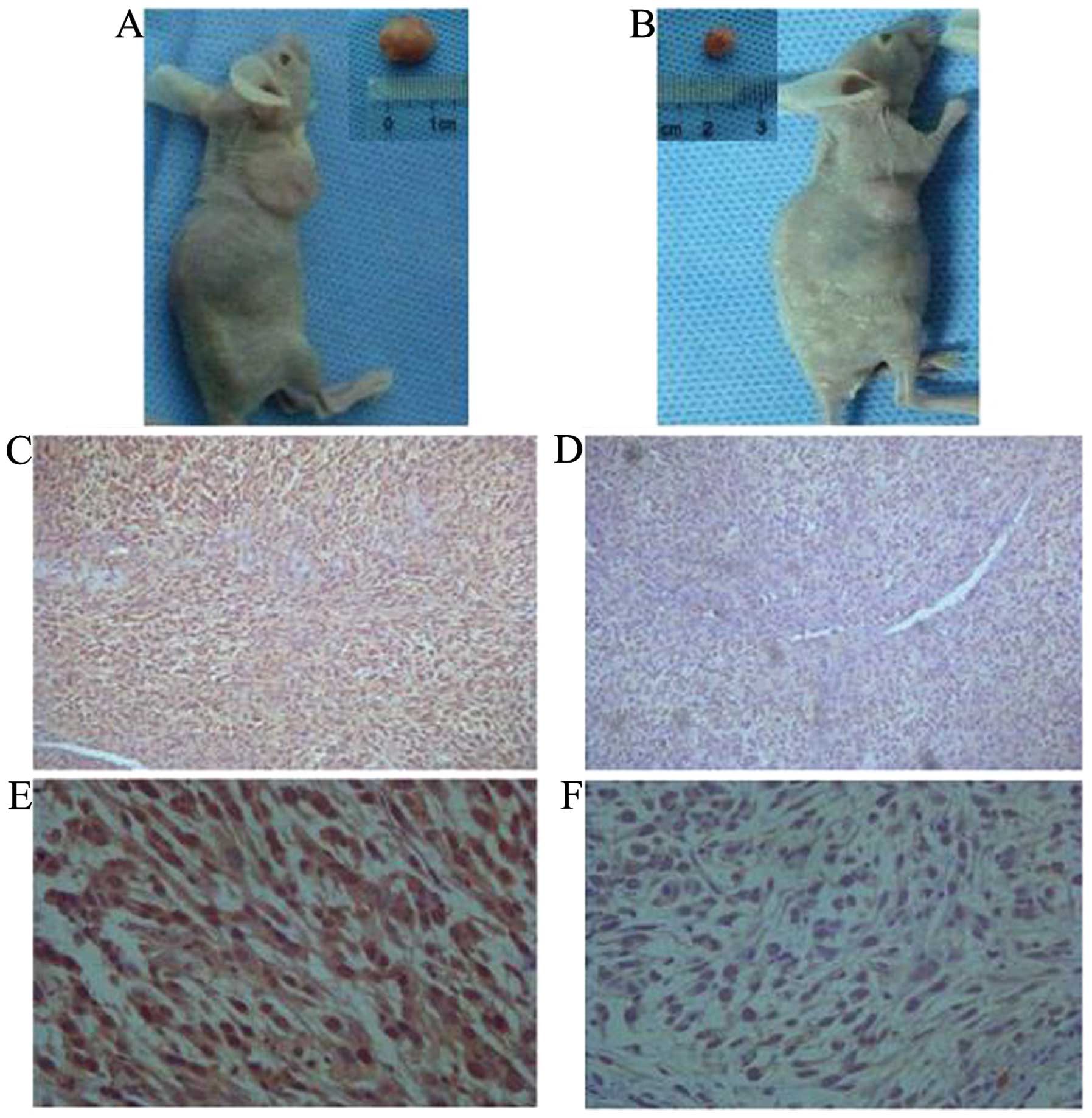
develop similar tumors (Figs. 3 and
4A and B). H&E staining
indicated that the ALDH+ and ALDH−-induced
tumors have histological differences. Furthermore,
ALDH+-induced tumors had a higher ALDH1 expression as
determined by immunostaining (Fig.
4C–F). These results suggested that ALDH1 may be the gene
responsible for the selected ALDH enzymatic activity.
 | Table IITumor-initiating capacity of
ALDH+ and ALDH− populations. |
Table II
Tumor-initiating capacity of
ALDH+ and ALDH− populations.
| Populations | Cell planting
|
|---|
|
1×103 |
5×103 |
5×104 |
5×105 |
5×106 |
|---|
|
ALDH+ | 1/5 | 2/5 | 4/5 | 5/5 | 5/5 |
|
ALDH− | 0/5 | 0/5 | 1/5 | 3/5 | 5/5 |
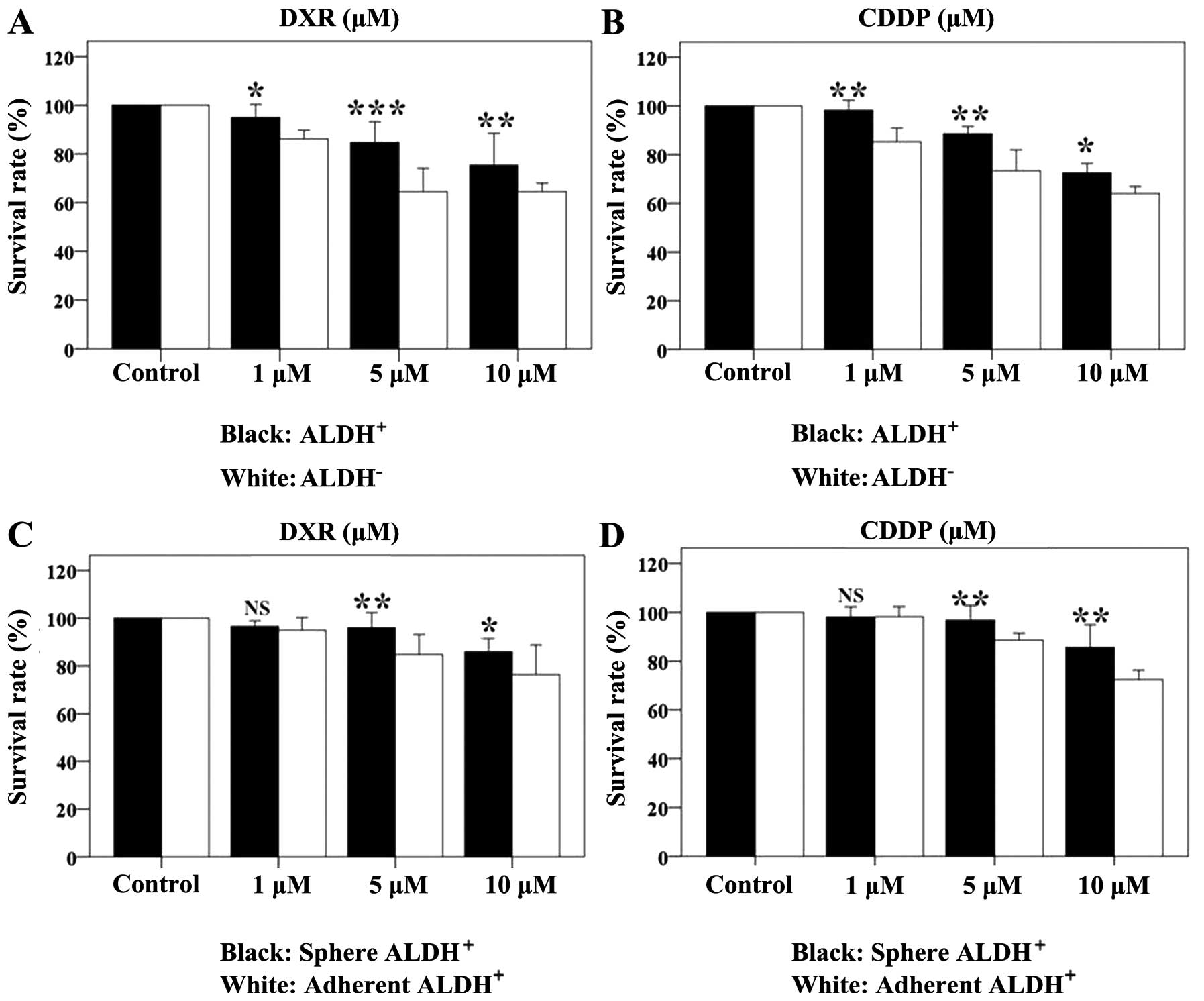
Drug efficacy on cell monolayers and
spheres
The results showed DXR and CDDP have dose-dependent
effects on cell survival. The ALDH+ and ALDH−
cell survival rate was determined after 48-h drug treatments
(Table III and IV, Fig.
5). DXR and CDDP killed the two cell populations; however, the
ALDH+ cells were more resistant to the drugs than the
ALDH− cells. In order to further validate the resistance
of the ALDH+ cells, we compared the drug response of
spheroid and adherent cells. The rate of growth inhibition of the
ALDH+ spheroids was slightly higher than that of
adherent cells in response to the two drugs (Table IV, Fig.
5C and D).
 | Table IIIALDH+ and ALDH−
cell survival rates (%) after 48-h treatment with DXR or CDDP. |
Table III
ALDH+ and ALDH−
cell survival rates (%) after 48-h treatment with DXR or CDDP.
| DXR
| CDDP
|
|---|
| 1 μM | 5 μM | 10 μM | 1 μM | 5 μM | 10 μM |
|---|
|
ALDH+ | 94.92±2.16 | 84.69±3.41 | 76.34±4.99 | 98.19±1.67 | 88.57±1.17 | 72.40±1.60 |
|
ALDH− | 86.18±1.39 | 64.56±3.83 | 64.56±1.39 | 85.25±2.26 | 73.31±3.48 | 64.08±1.14 |
| F-value | 11.86 | 89.68 | 49.78 | 52.44 | 51.88 | 11.15 |
| P-value | 0.026 | 0.001 | 0.002 | 0.002 | 0.002 | 0.029 |
 | Table IVSpherical and adherent cell survival
rates (%) after 48-h treatment with DXR or CDDP. |
Table IV
Spherical and adherent cell survival
rates (%) after 48-h treatment with DXR or CDDP.
| DXR
| CDDP
|
|---|
| 1 μM | 5 μM | 10 μM | 1 μM | 5 μM | 10 μM |
|---|
| Sphere | 96.4±0.78 | 95.95±2.58 | 85.90±2.22 | 98.11±1.68 | 96.80±2.43 | 85.58±3.77 |
| Adhere | 94.92±2.16 | 84.69±3.41 | 76.34±4.99 | 98.19±1.67 | 88.57±1.17 | 72.40±1.60 |
| F-value | 1.035 | 20.83 | 9.019 | 0.000 | 28.00 | 31.11 |
| P-value | 0.310 | 0.010 | 0.039 | 0.960 | 0.006 | 0.005 |
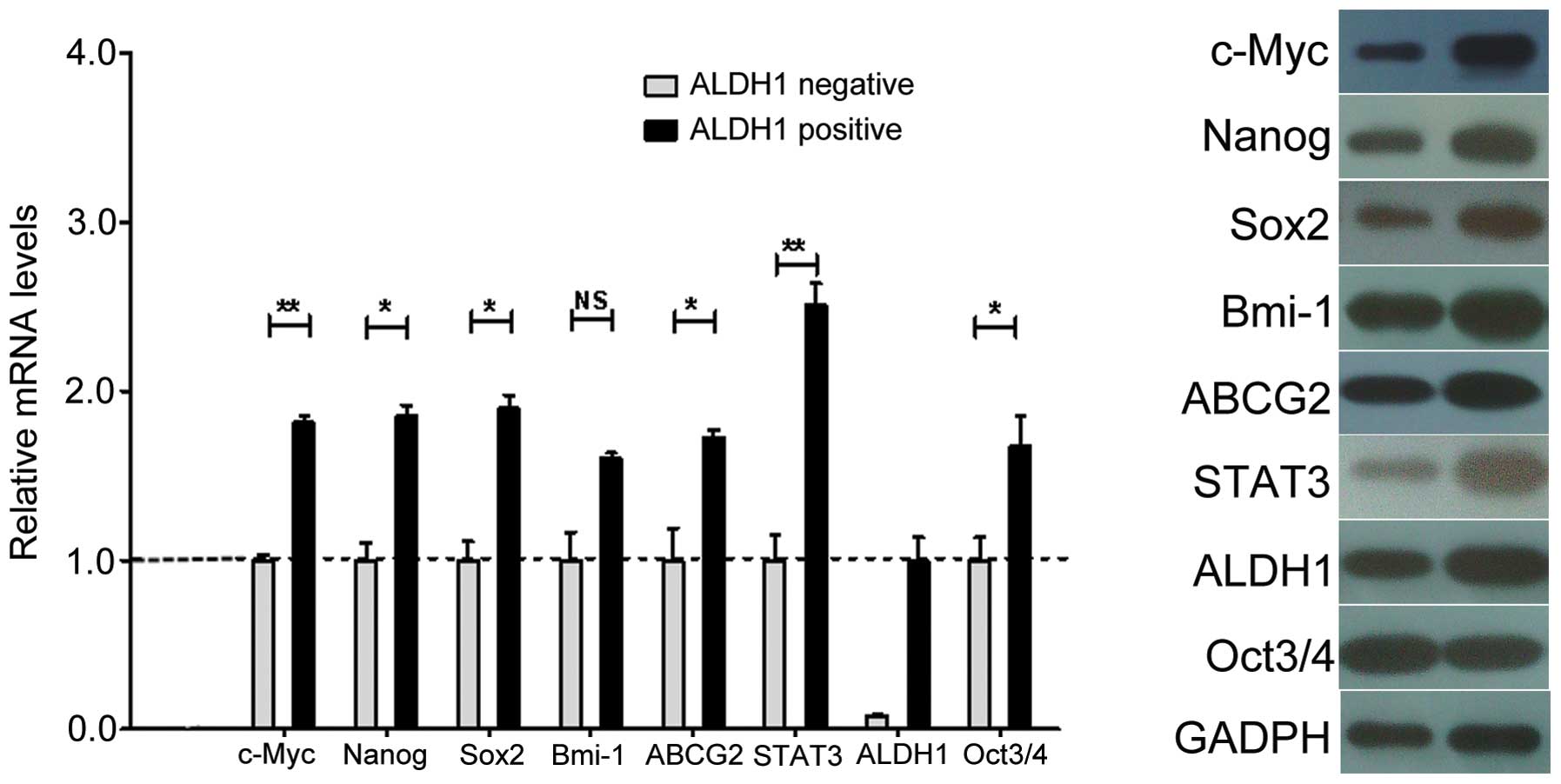
qPCR
Stemness and drug transporter genes are
characteristic of CSCs. To determine whether the NMFH-1
subpopulations shared these characteristics, we measured the
expression of the c-Myc, Bmi-1, Sox2,
Nanog, Oct3/4, STAT3, ABCG2 and
ALDH1 genes (19–21). The ALDH+ population had
increased levels of ALDH1 gene expression. Therefore the CSC
population that we isolated can also be referred to as
ALDH1+. We found that Bmi-1 expression was not
significantly different between the ALDH+ and
ALDH− cells. However, the expression of c-Myc,
STAT3, Sox2, Nanog, Oct3/4, and
ABCG2 was significantly increased in the ALDH+
cells as measured by mRNA transcript levels (Fig. 6A). However, the expression of Oct3/4
protein was not higher in the ALDH+ cells compared with
the ALDH− cells (Fig.
6B). The transcription and protein expression of the ABCG2 drug
transporter was significantly increased in the ALDH+
cells compared with the ALDH− cells. Thus, these data
demonstrated that the NMFH-1 cells have an ALDH+
subpopulation that expresses genes highly associated with stem
cells.
Discussion
Tumors are difficult to cure due to the existence of
CSCs, which have the potential for self-renewal, unlimited
proliferation, and differentiation into tumor cells (1,2).
Previous findings confirm that CSCs exist in many tumor tissues.
However, the isolation of CSCs to develop improved cancer
treatments in the future remains to be investigated.
In 1996, the identification of a side population
(SP) of cells was proposed as a new method for stem cell
separation. The SP, defined by Hoechst 33342 dye exclusion, was
identified as a distinct subset of cells. These cells have
subsequently been isolated and designated CSCs because they possess
stemness characteristics and are responsible for tumorigenesis in
several cancer types (22–25).
In 2003, Al-Hajj et al (26) isolated CSCs (~2%) from breast cancer
specimens with the cell surface markers
CD24−CD44+Lin−, and verified that
these cells were highly tumorigenic. Another series of cell surface
markers, such as CD133, CD90, CD44, CD34, and CD38 have also been
used to isolate CSCs (27–31). These specific surface markers are
mostly present in solid tumors. However, stem cells from different
tissues may express unique markers.
ALDH1 has recently been identified as a marker for
CSCs in humans, because many types of stem cells express ALDH1 at
high levels. This observation has been confirmed in breast stem
cells, HSCs, neural stem cells, prostate, colon and lung CSCs
(6,7,32–36).
As a functional protease, the gene is more common than the cell
surface markers. Confirmation that all the stem cells express high
levels of ALDH1+ may provide a reliable method for the
isolation of the CSCs. Our results show that 1.74% of the NMFH-1
cells are ALDH+. We also found that the survival rate
following the freeze-thaw of the ALDH+ cells was
significantly higher than that in the ALDH− cells.
However, subsequent generations of these cells did not have
significant growth defects. It is possible that the low temperature
had a specific-temporary effect on the growth of the
ALDH− cells.
The results showed that the ALDH+ NMFH-1
cells are more capable of forming spheres when cultured in
serum-free medium than the ALDH− cells, and these cells
can form spheres a second time. The ALDH− cells required
longer time periods and formed smaller spheres. These spheres may
have been formed by the CSCs of other subpopulations marked by
CD133, CD44 or other markers. It is possible that ALDH selection
could isolate most of the stem cells but not all of them; however,
this remains to be demonstrated. The ability of the
ALDH+ cells to form tumors in nude mice is very
apparent. Briefly, 1×103 ALDH+ cells grown in
nude mice subcutaneously formed a tumor, whereas 5×104
ALDH− cells did not form tumor in nude mice.
Furthermore, the ALDH+ cells formed larger tumors than
the ALDH− cells during the same time-period. Previous
studies have indicated that 500 ALDH+ cells can form
tumors but 5,000 ALDH− cells are required to form tumors
(6). Our experiments required more
cells which may be an indication of differences in the laboratory
conditions, personnel operation, or experimental design.
Immunohistochemistry confirmed that the tumors developed from the
ALDH+ cells expressed high levels of the ALDH1 protein
in mice. By contrast, low levels of the ALDH1 protein were
expressed in tumors developed from the ALDH− cells.
Thus, it can be hypothesized that the ALDH1+ cells
contribute to tumor formation, which is consistent with these cells
being CSCs. In the present study, ALDH1 expression was maintained
even after several rounds of division in vivo. However, in
ordinary tumor cells, the content of ALDH1 is very limited.
Survival competition of the ALDH1− cells may account for
this phenomenon by secreting some factors that limit the
ALDH1+ cell proliferation. However, this remains an open
area of research.
We compared the expression of genes that play a
prominent role in stem cell maintenance, self-renewal, and nuclear
reprogramming. These included c-Myc, Bmi-1,
Sox2, Nanog, Oct3/4 and STAT3 in
ALDH+ and ALDH− cells. If these cells
expressed these genes at high levels, they could not all be defined
as CSCs. We used ALDH enzymatic activity as the marker for
isolation and we detected ALDH1 gene expression in both the
ALDH+ and ALDH− cells. qPCR assays showed
that the expression of c-Myc and STAT3 was
significantly different between populations (P<0.01). c-Myc
regulates the G0-G1 cell cycle transition and promotes cell
division. It has also been shown to control infinite rounds of
proliferation and the development of tumor formation. Many growth
factors can stimulate fibroblast cells, which can lead to an
enhanced c-Myc expression. Bmi-1 emerged as a Myc-cooperating
oncogene (37–39). STAT3 is a signal transduction factor
and an important member of a family of activating factors. The STAT
signaling pathways are closely associated with cell proliferation,
differentiation and apoptosis. Activation of the pathway can lead
to abnormal cell proliferation and malignant transformation
(35,40). The expression of Sox2, Nanog, and
Oct3/4 was significantly different between the ALDH+ and
ALDH− populations (P<0.05). Sox2 and Oct3/4 can
cooperate to control fibroblast growth factor 4 (FGF4). FGF4 is a
signaling molecule that plays an important role in embryonic
development, and the FGF4 gene has a specific enhancer
element in the 3′UTR (41–43). Nanog encodes a recently identified
divergent homeoprotein that controls cell self-renewal (44).
In the drug resistance experiment, we found that the
NMFH-1 ALDH+ cells were more resistant than the
ALDH− cells. Moreover, the ALDH+-derived
spheres showed greater resistance than the ALDH+
adherent cells. In clinical practice, MFH is difficult to cure and
is not sensitive to these drugs. Comprehensive surgery, radiation,
and chemotherapy are known treatment methods used to improve the
resection and survival rates, and reduce the local recurrence rate
of MFH (45,46). ABCG2 expression is reported to
significantly contribute to the CSC phenotype, strongly correlate
with drug resistance, and indicate a poor clinical outcome
(47,48). The mRNA expression of the
ABCG2 gene between populations had a significant difference,
as shown by the RT-PCR results. In the ALDH+ cells, the
ABCG2 protein expression was higher than that in the
ALDH− cells, which potentially accounts for
ALDH+ cell drug resistance.
In conclusion, our study is the first to
successfully isolate the ALDH+ subpopulation from the
NMFH-1 cell line. The experiments also show that the
ALDH+ subpopulation exhibits several characteristic CSC
properties, including high clonogenicity and self-renewal,
increased chemotherapeutic drug resistance, elevated expression of
stemness and drug transporter genes and high tumorigenic potential.
The above results show that ALDH1 can be used as a marker for
isolation of the CSCs from the NMFH-1 cell line. We hypothesize
that ALDH1 may be used as a biomarker for stem cell specificity and
applied to other tumors. ALDH1 expression may also play a future
role in the development of clinical treatments.
Acknowledgments
This study was supported by a grant from the
National Natural Science Foundation of China (no. 81072192). We
would like to thank Dr Akira Ogose (Division of Orthopedic Surgery,
Niigata University Graduate School of Medical and Dental Sciences,
Niigata, Japan) for providing the NMFH-1 cell line.
References
|
1
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molofsky AV, Pardal R and Morrison SJ:
Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell
Biol. 16:700–707. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fabrizi E, di Martino S, Pelacchi F and
Ricci-Vitiani L: Therapeutic implications of colon cancer stem
cells. World J Gastroenterol. 16:3871–3877. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Piscaglia AC: Stem cells, a two-edged
sword: risks and potentials of regenerative medicine. World J
Gastroenterol. 14:4273–4279. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di J, Duiveman-de Boer T, Figdor CG and
Torensma R: Aiming to immune elimination of ovarian cancer stem
cells. World J Stem Cells. 5:149–162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ginestier C, Hur MH, Charafe-Jauffret E,
et al: ALDH1 is a marker of normal and malignant human mammary stem
cells and a predictor of poor clinical outcome. Cell Stem Cell.
1:555–567. 2007. View Article : Google Scholar
|
|
7
|
Jiang F, Qiu Q, Khanna A, et al: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Purton LE: Roles of retinoids and retinoic
acid receptors in the regulation of hematopoietic stem cell
self-renewal and differentiation. PPAR Res. 2007:879342007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Purton LE, Bernstein ID and Collins SJ:
All-trans retinoic acid delays the differentiation of primitive
hematopoietic precursors (lin-c-kit+Sca-1(+)) while enhancing the
terminal maturation of committed granulocyte/monocyte progenitors.
Blood. 94:483–495. 1999.PubMed/NCBI
|
|
10
|
Jones RJ, Barber JP, Vala MS, Collector
MI, Kaufmann SH, Ludeman SM, Colvin OM and Hilton J: Assessment of
aldehyde dehydrogenase in viable cells. Blood. 85:2742–2746.
1995.PubMed/NCBI
|
|
11
|
Storms RW, Trujillo AP, Springer JB, Shah
L, Colvin OM, Ludeman SM and Smith C: Isolation of primitive human
hematopoietic progenitors on the basis of aldehyde dehydrogenase
activity. Proc Natl Acad Sci USA. 96:9118–9123. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hess DA, Meyerrose TE, Wirthlin L, Craft
TP, Herrbrich PE, Creer MH and Nolta JA: Functional
characterization of highly purified human hematopoietic
repopulating cells isolated according to aldehyde dehydrogenase
activity. Blood. 104:1648–1655. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Corpron CA, Black CT, Raney RB, Pollock
RE, Lally KP and Andrassy RJ: Malignant fibrous histiocytoma in
children. J Pediatr Surg. 31:1080–1083. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanzaki T, Kitajima S and Suzumori K:
Biological behavior of cloned cells of human malignant fibrous
histiocytoma in vivo and in vitro. Cancer Res. 51:2133–2137.
1991.PubMed/NCBI
|
|
15
|
Schmidt H, Körber S, Hinze R, Taubert H,
Meye A, Würl P, Holzhausen HJ, Dralle H and Rath FW: Cytogenetic
characterization of ten malignant fibrous histiocytomas. Cancer
Genet Cytogenet. 100:134–142. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meloni-Ehrig AM, Chen Z, Guan XY,
Notohamiprodjo M, Shepard RR, Spanier SS, Trent JM and Sandberg AA:
Identification of a ring chromosome in a myxoid malignant fibrous
histiocytoma with chromosome microdissection and fluorescence in
situ hybridization. Cancer Genet Cytogenet. 109:81–85. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawashima H, Ogose A, Gu W, et al:
Establishment and characterization of a novel myxofibrosarcoma cell
line. Cancer Genet Cytogenet. 161:28–35. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gibbs CP, Kukekov VG, Reith JD,
Tchigrinova O, Suslov ON, Scott EW, Ghivizzani SC, Ignatova TN and
Steindler DA: Stem-like cells in bone sarcomas: implications for
tumorigenesis. Neoplasia. 7:967–976. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okita K, Ichisaka T and Yamanaka S:
Generation of germline-competent induced pluripotent stem cells.
Nature. 448:313–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wernig M, Meissner A, Foreman R, Brambrink
T, Ku M, Hochedlinger K, Bernstein BE and Jaenisch R: In vitro
reprogramming of fibroblasts into a pluripotent ES-cell-like state.
Nature. 448:318–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lathia JD: Cancer stem cells: moving past
the controversy. CNS Oncol. 2:465–467. 2013. View Article : Google Scholar
|
|
23
|
Haraguchi N, Utsunomiya T, Inoue H, Tanaka
F, Mimori K, Barnard GF and Mori M: Characterization of a side
population of cancer cells from human gastrointestinal system. Stem
Cells. 24:506–513. 2006. View Article : Google Scholar
|
|
24
|
Chiba T, Kita K, Zheng YW, Yokosuka O,
Saisho H, Iwama A, Nakauchi H and Taniguchi H: Side population
purified from hepatocellular carcinoma cells harbors cancer stem
cell-like properties. Hepatology. 44:240–251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szotek PP, Pieretti-Vanmarcke R, Masiakos
PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F,
Maclaughlin DT and Donahoe PK: Ovarian cancer side population
defines cells with stem cell-like characteristics and mullerian
inhibiting substance responsiveness. Proc Natl Acad Sci USA.
103:11154–11159. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514. 2008.
View Article : Google Scholar
|
|
28
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of
CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Armstrong L, Stojkovic M, Dimmick I, Ahmad
S, Stojkovic P, Hole N and Lako M: Phenotypic characterization of
murine primitive hematopoietic progenitor cells isolated on basis
of aldehyde dehydrogenase activity. Stem Cells. 22:1142–1151. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Thant AA, Machida K, Ichigotani
Y, Naito Y, Hiraiwa Y, Senga T, Sohara Y, Matsuda S and Hamaguchi
M: Hyaluronan-CD44s signaling regulates matrix metalloproteinase-2
secretion in a human lung carcinoma cell line QG90. Cancer Res.
62:3962–3965. 2002.PubMed/NCBI
|
|
32
|
Hess DA, Wirthlin L, Craft TP, Herrbrich
PE, Hohm SA, Lahey R, Eades WC, Creer MH and Nolta JA: Selection
based on CD133 and high aldehyde dehydrogenase activity isolates
long-term reconstituting human hematopoietic stem cells. Blood.
107:2162–2169. 2006. View Article : Google Scholar
|
|
33
|
Seigel GM, Campbell LM, Narayan M and
Gonzalez-Fernandez F: Cancer stem cell characteristics in
retinoblastoma. Mol Vis. 11:729–737. 2005.PubMed/NCBI
|
|
34
|
Ivashkiv LB: Jak-STAT signaling pathways
in cells of the immune system. Rev Immunogenet. 2:220–230.
2000.
|
|
35
|
Kim H, Lapointe J, Kaygusuz G, Ong DE, Li
C, van de Rijn M, Brooks JD and Pollack JR: The retinoic acid
synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate
cancer. Cancer Res. 65:8118–8124. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang EH, Hynes MJ, Zhang T, Ginestier C,
Dontu G, Appelman H, Fields JZ, Wicha MS and Boman BM: Aldehyde
dehydrogenase 1 is a marker for normal and malignant human colonic
stem cells (SC) and tracks SC overpopulation during colon
tumorigenesis. Cancer Res. 69:3382–3389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brunk BP, Martin EC and Adler PN:
Drosophila genes posterior sex combs and suppressor two of zeste
encode proteins with homology to the murine bmi-1 oncogene. Nature.
353:351–353. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Iwama A, Oguro H, Negishi M, et al:
Enhanced self-renewal of hematopoietic stem cells mediated by the
polycomb gene product Bmi-1. Immunity. 21:843–851. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lessard J and Sauvageau G: Bmi-1
determines the proliferative capacity of normal and leukaemic stem
cells. Nature. 423:255–260. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ning ZQ, Li J, McGuinness M and Arceci RJ:
STAT3 activation is required for Asp(816) mutant c-Kit induced
tumorigenicity. Oncogene. 20:4528–4536. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maucksch C, Jones KS and Connor B: Concise
review: the involvement of SOX2 in direct reprogramming of induced
neural stem/precursor cells. Stem Cells Transl Med. 2:579–583.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dailey L, Yuan H and Basilico C:
Interaction between a novel F9-specific factor and octamer-binding
proteins is required for cell-type-restricted activity of the
fibroblast growth factor 4 enhancer. Mol Cell Biol. 14:7758–7769.
1994.PubMed/NCBI
|
|
43
|
Ambrosetti DC, Basilico C and Dailey L:
Synergistic activation of the fibroblast growth factor 4 enhancer
by Sox2 and Oct-3 depends on protein-protein interactions
facilitated by a specific spatial arrangement of factor binding
sites. Mol Cell Biol. 17:6321–6329. 1997.PubMed/NCBI
|
|
44
|
Mitsui K, Tokuzawa Y, Itoh H, Segawa K,
Murakami M, Takahashi K, Maruyama M, Maeda M and Yamanaka S: The
homeoprotein Nanog is required for maintenance of pluripotency in
mouse epiblast and ES cells. Cell. 113:631–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Le Doussal V, Coindre JM, Leroux A, Hacene
K, Terrier P, Bui NB, Bonichon F, Collin F, Mandard AM and Contesso
G: Prognostic factors for patients with localized primary malignant
fibrous histiocytoma: a multicenter study of 216 patients with
multivariate analysis. Cancer. 77:1823–1830. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ágoston P, Kliton J, Mátrai Z and Polgár
C: Radiotherapy of soft tissue sarcomas of the extremities and
superficial trunk. Magy Onkol. 58:65–76. 2014.In Hungarian.
|
|
47
|
Castillo V, Valenzuela R, Huidobro C,
Contreras HR and Castellon EA: Functional characteristics of cancer
stem cells and their role in drug resistance of prostate cancer.
Int J Oncol. 45:985–994. 2014.PubMed/NCBI
|
|
48
|
Ding XW, Wu JH and Jiang CP: ABCG2: a
potential marker of stem cells and novel target in stem cell and
cancer therapy. Life Sci. 86:631–637. 2010. View Article : Google Scholar : PubMed/NCBI
|