Cancer is considered a terrible disease that leads
to a global health threat. Although significant improvements have
been made in the management of cancer as well as the comprehension
of the molecular mechanisms of neoplasm pathogenesis and
progression, cancer remains a common disorder worldwide, accounting
for 12.7 million new cancer cases and 7.6 million cancer deaths in
2008, worldwide (1,2). Previous findings have shown that
tumorigenesis in humans is a multi-step process foundational with
mutations which can activate oncogenes and inactivate
tumor-suppressor genes. The genetic alterations can result in
changes of the proteome, and these changes eventually drive the
malignant biological behavior through complex signaling pathways
(3–9).
The ubiquitin-proteasome system (UPS), which is
responsible for the degradation of >80% of cell proteins, is the
main proteolytic mechanisms involved in eukaryocytes (3,4). Most
of the proteins involved in cell cycle progression, proliferation
and apoptosis are regulated by the UPS (5). Dysregulation of the UPS may contribute
to tumor progression, drug resistance and altered immune
surveillance (5). Previous studies
have focused on the relationship between epithelial-mesenchymal
transition (EMT) signal transduction pathways and UPS, and found
that many signal transducers and transcription factors involved in
EMT are regulated by ubiquitination and the UPS (6). It is well-known that the UPS consists
of three classes of enzymes: the ubiquitin-activating enzyme (E1),
the ubiquitin-conjugating enzyme (E2) and ubiquitin ligases (E3).
In most cases, the three enzymes together with 26S proteasome drive
target substrate degradation through a series of catalytic
processes (5,7). In the human genome there are two E1s,
~30–40 E2s and >600 E3 ligases (8). It should be emphasized that during the
ubiquitination, E3 mediates the transfer of ubiquitin from E2 to
the substrate protein. Thus, it has a central role in the substrate
decision (9).
The SCF complexes belong to the cullin-RING ligase
family and are the largest family of E3 ligases comprising four
subunits: S-phase kinase-associated protein 1 (SKP1), which is
responsible for recruiting the variable F-box protein; cullin 1
(CUL1), which provides a rigid scaffold connecting SKP1 and RBX1 on
opposite ends; RING box 1 (RBX1; also known as ROC1 or HRT1), which
serves as an interface for E2 ubiquitin-conjugating enzymes; and a
member of the F-box protein family, which functions as a
substrate-recognition component (10,11).
F-box and WD repeat domain-containing 7 (Fbxw7; also known as Fbw7,
hCdc4, hAGO and Sel10) is an evolutionarily conserved protein
belonging to the F-box family. It was first identified as Cdc4 in
budding yeast in 1973 (12). Sel10
was subsequently identified in Caenorhabditis elegans as a
negative regulator of Lin-12 (homology of Notch) (13). In parallel, archipelago, a
gene in Drosophila melanogaster encoding a protein
containing an F-box domain and seven tandem WD repeats (AGO)
domain, was identified (14). From
those studies, the human homologue was ascertained and was
designated as Fbxw7 or Fbw7.
Fbxw7 is thought to be a tumor suppressor involved
in cell growth, proliferation, differentiation and survival
(15). Fbxw7 has been found to be
inactivated by mutation in various human cancer types (16–18).
Fbxw7 has an unusual mutation spectrum whereby biallelic, simple
loss-of-function mutations are rare. Instead, most mutations are
monoallelic missense changes involving specific arginine residues
at β-sheet propellor tips that allow the Fbxw7 protein to recognize
its substrates (19). Inactivation
of the Fbxw7 protein is associated with the deregulation of several
well-known oncoproteins with significant capabilities in pathways
that manage cell division and growth, including cyclin E (20), c-Myc (21), c-Jun (22), Notch (23), Mcl-1 (24) and mammalian target of rapamycin
(mTOR) (25) (Table I).
Therefore, the dysregulation of Fbxw7-mediated
proteasome degradation is likely involved in many signaling
pathways which play important roles in human cancers. Among these
oncogenic Fbxw7 substrates, cyclin E and c-Myc may be the most
clearly investigated oncoproteins that have certain contributions
to Fbxw7-associated cancers. In this review, we discuss the two
major signaling pathways impacted by Fbxw7 to show how
Fbxw7-related signaling pathways are regulated in cancer. In
addition, since few studies have focused on the relationship
between hepatocellular carcinoma (HCC) and Fbxw7, we assess
molecular mechanisms by which Fbxw7 exerts antitumor activity in
HCC.
Fbxw7, which shares the WD40 repeats structure (a
substrate interaction domain used to classify F-box proteins), is a
well-studied member of the F-box family (54). The human FBXW7 gene consists
of 4 introns and 13 exons and its gene locus maps to chromosome
region 4q32, which is commonly deleted in many types of human
malignancies (17,55). The FBXW7 gene encodes the
Fbxw7α, Fbxw7β and Fbxw7γ protein isoforms, which have distinct
subcellular localizations, with Fbxw7α mainly localizing to
nucleoplasm, Fbxw7β to cytoplasm and Fbxw7γ to nucleolus (56). The three isoforms also seem to have
tissue specificity with the α-form being found to be extensively
expressed in human tissues, while the β-form and/or γ-form are
present at lower levels, except in skeletal muscle, brain and to a
lesser degree, heart (17).
In addition to the N-terminal region which contains
dominant signals for the subcellular localization, each isoform of
Fbxw7 shares conserved interaction domains in the C-terminal
region: the eight WD40 repeats that determine target specificity;
the F-box that recruits the SKP1 of the SCF complex; and the D
domain that dimerize the SCFFbxw7, which allows it to
target substrates with low-affinity Cdc4 phosphodegron (CPDs)
(15). All these domains are
essential for the degradation of its substrates.
Although the three Fbxw7 isoforms share many
identical functional domains, recent studies have identified that
each isoform has its special function. The α-form is most abundant
and accounts for degradation of most tested substrates. Fbxw7β is
proved to reside in the endoplasmic reticulum membrane and protects
cells from oxidative stress (57).
Moreover, Fbxw7α and Fbxw7β are found to play an opposite role in
their substrates. For example, PGC-1α, a transcriptional
coactivator with broad effects on cellular energy metabolism, is
found to have a different fate under different Fbxw7 isoforms
(52). Fbw7β reduces cellular
PGC-1α via ubiquitin-mediated degradation, whereas Fbw7α increases
cellular PGC-1α via ubiquitin-mediated stabilization (52).
Evidence suggests that substrates of Fbxw7 are
polyubiquitinated in a GSK3-dependent manner by SCFFbxw7
(22,33,58).
Glycogen synthase kinase 3 (GSK3), firstly identified in 1980, is a
constitutively active and ubiquitously expressed serine/threonine
kinase (59,60). Human cells contain two GSK3
isoforms, known as GSK3α and GSK3β, which are highly similar with
respect to sequence (share 97% amino acid sequence within their
catalytic domains) and function (61). The activation of GSK3 is dominated
by phosphorylation on Ser-21 of GSK3α and Ser-9 of GSK3β (62). It is well established that these
phosphorylations are regulated via PI3K/Akt pathway (63). In most cases, inactivation of the
PI3K/Akt pathway leads to dephosphorylation of GSK3, which results
in the activation of GSK3. The activated GSK3 subsequently
phosphorylates the CPDs of the substrates that are primed by
phosphorylation at position +4 of the CPD by a yet to-be-identified
kinase (in the cyclin E case, which is Cdk2) (33,64).
The CPDs of substrates are recognized and interacted with the eight
WD40 repeats of Fbxw7 for ubiquitin-mediated proteolytic
degradation (65).
CPD is a phosphodegron motif existing in the
substrates of Fbxw7. The phosphorylation of CPD by GSK3 is crucial
for the interaction between Fbxw7 and its substrate. Mutation of
critical residues within the CPD lead to stabilization of
substrates, such as c-Myc and has been observed in many human
cancers (66). Data gathering from
Fbxw7 substrates have ascertained the conserved CDP sequence as
ΦXΦΦΦ-T/S-PPX-S/T/E, with Φ standing for a hydrophobic residue and
X for any amino acid (65,67). The T/S residue can be phosphorylated
by GSK3 and the phosphorylation of S/T/E residue serves as a
priming signal for GSK3 phosphorylation. Moreover, studies also
found that some substrates have more than one CPD. For example,
studies have revealed that cyclin E has two CPDs, with one located
in the T380 and the other one being centered at ~T62 (68,69).
The two CPDs of cyclin E were essential for efficiency of Fbxw7
binding, in response to different signaling pathways (70).
Over the past decade, a number of studies have
contributed to the understanding of Fbxw7 molecular mechanisms in
human cancers. Several molecules that play an essential role in
tumor pathogenesis and progression have been identified to be
regulated by Fbxw7, and are now recognized as potential therapeutic
targets. In the present review, we focus on the mechanism by which
cyclin E and c-Myc are regulated by Fbxw7.
By binding to its kinase partner Cdk2, cyclin E
regulates the cell cycle by promoting the G1-S transition (71). Deregulation of cyclin E causes
genomic instability and is thought to directly contribute to cell
transformation and tumorigenesis (72). Cyclin E, containing two CPDs that
are phosphorylated by GSK3 and autophosphorylated by Cdk2,
respectively, is the most well-studied substrate of Fbxw7 (69). Cyclin E has gained much attention as
a key mediator for the tumor-suppressor ability of Fbxw7.
Specifically, Fbxw7 tightly regulates the abundance of cyclin E
through many molecular mechanisms.
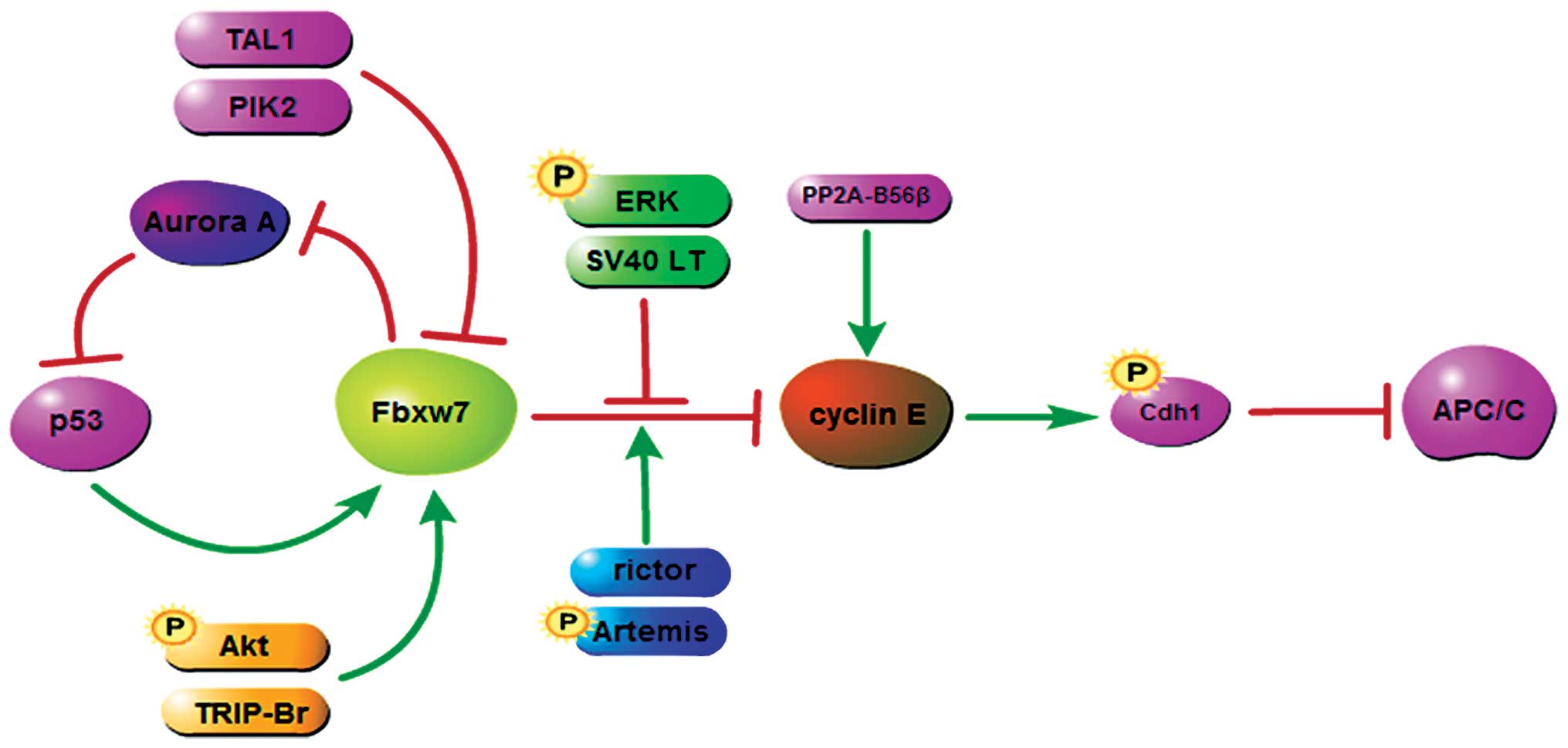
p53 has a complicated and incompletely understood
interplay between Fbxw7 and cyclin E. Previous observations
revealed that the p53-p21 pathway, which is induced by excess
cyclin E, suppresses cyclin E kinase activity and this pathway
cooperates with SCFFbxw7 to suppress cyclin E-induced
genome instability (73,74). Moreover, when using a
cDNA-microarray system, p53 protein is found to be a
transcriptional activator of FBXW7b (75). p53 arrests cell cycle progression at
G0-G1 by inducing the Fbxw7β-mediated downregulation of cyclin E
expression (75). Similarly,
another group provided results showing that, p53 regulates the
levels of cyclin E protein to impose a G1-S block through the
activation of ago in Drosophila (76). However, previous findings have also
shown that, Fbxw7β is not necessary for the degradation of cyclin E
in mammals as mentioned below. Fbxw7 was eventually proven to act
as an upstream of p53 by inducing Aurora A degradation (77). Aurora A phosphorylates p53 at
S315/215 and thus reduces p53 levels and transcriptional activity
(77) (Fig. 1). However, more studies are needed
to clarify these paradoxical and complicated relationships.
Some molecules influence the process whereby Fbxw7
modulates cyclin E protein degradation, for instance rictor
(78), Artemis (79) and SV40 large T (LT) (80) (Fig.
1). Although rictor is usually identified as a binding partner
of mTOR, numerous complexes containing rictor have been recognized
and shown to be mTOR-independent (81). Fbxw7α was recently shown to form a
complex with rictor. The rictor/Fbxw7 complex functions as an E3
ligase complex, and promotes the degradation of cyclin E in an
mTOR-independent manner (78).
Artemis, a member of the SNM1 gene family, is a known
phosphorylation target of ATM, ATR and DNA-PKcs in response to
various types of genotoxic stress (82). In response to UV irradiation,
Artemis is phosphorylated at S516 and S645 by the ATR kinase. The
phosphorylated Artemis then interacts with Fbxw7α or Fbxw7γ and
induces strong ubiquitylation of cyclin E (79). Similarly, the knockdown of rictor by
shRNA and inactivation of Artemis by the mutation of S516/645 to
alanine can lead to a decreasing ubiquitination of cyclin E
(78,79). Contrary to rictor and Artemis, LT
negatively regulates the degradation of cyclin E by Fbxw7. LT is a
viral oncoprotein producing in cells infected with simian virus 40
(SV40) (83). SV40 LT protein binds
to a number of host cell proteins and disrupts their normal
functions. Welcker and and Clurman (80) found that LT also has a consensus CPD
in its C terminus that can be phosphorylated at T701, through which
LT functions as a competitive inhibitor of Fbxw7 and then
deregulates ubiquitination of cyclin E. The Ras/MAPK pathway is
also involved in the induction of cyclin E stability by altering
the physical interaction between Fbxw7 and cyclin E, but not by
altering cyclin E phosphorylation on any of its known regulatory
sites (84,85) (Fig.
1).
In addition to interacting with Fbxw7, some factors
may target cyclin E and disturb the interaction between Fbxw7 and
cyclin E. For example, the protein phosphatase PP2A-B55β targets
the N- and C-terminal phosphodegrons of cyclin E1 for
dephosphorylation, thereby protecting it from degradation mediated
by the SCFFbxw7 ubiquitin ligase (86) (Fig.
1). These data suggest the interaction between Fbxw7 and cyclin
E is tightly regulated by different regulators. However, as yet
unknown regulators remain to be identified in the future.
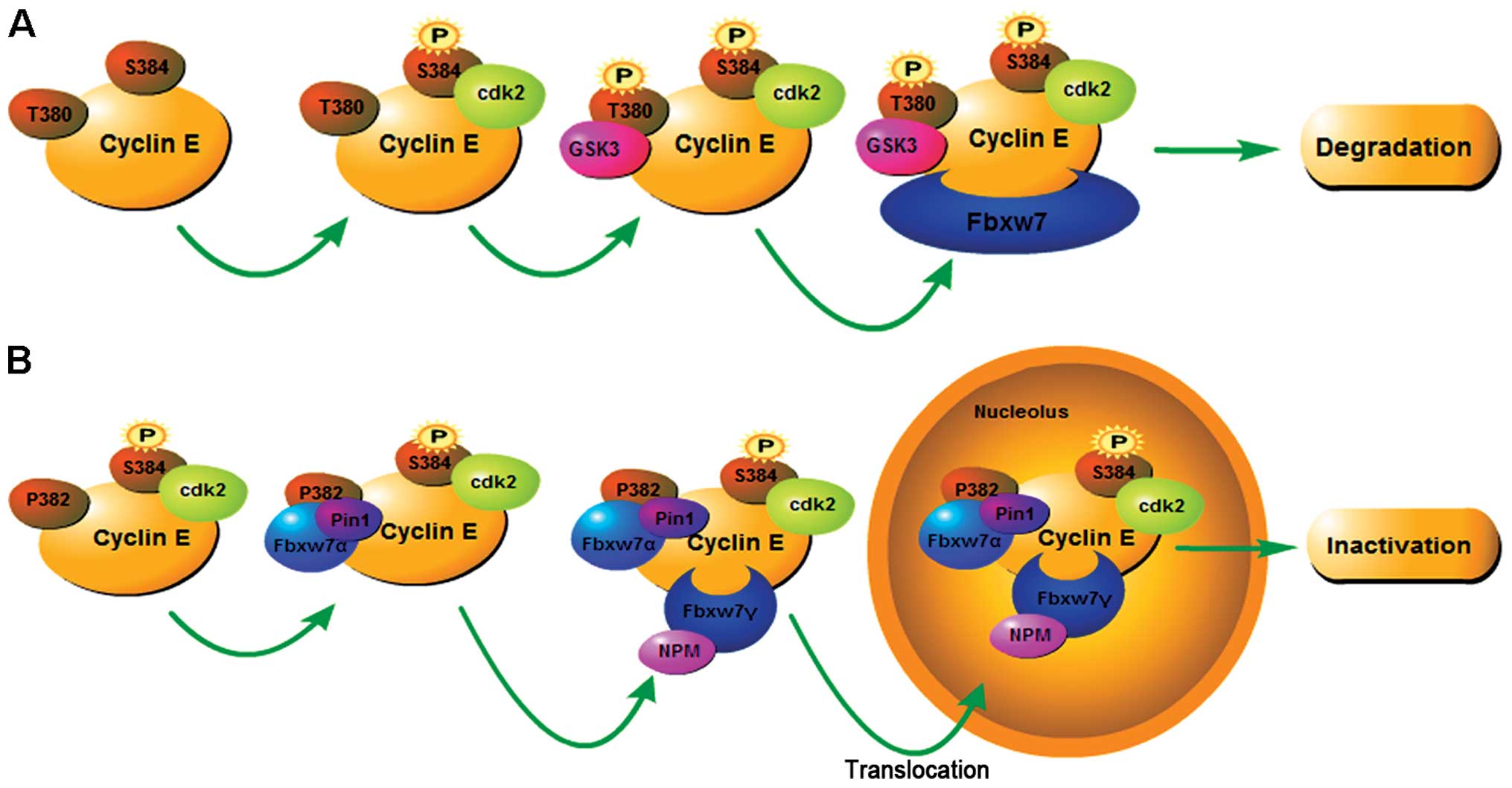
It has been demonstrated that ubiquitin-mediated
degradation of cyclin E in mammals can be divided into two ways:
one requires only Fbxw7α and the other requires Fbxw7α and Fbxw7γ,
although not Fbxw7β (56,87,88)
(Fig. 2). When the expression of
cyclin E is low, presumably normal, the inactivation of cyclin E
requires Fbxw7α and Fbxw7γ in a two-step manner. Firstly, cyclin E,
which is phosphorylated at S384 by Cdk2, interacts with prolyl
cis-trans isomerase Pin1 in conjunction with SCFFbxw7α,
carries out a noncanonical isomerization of a proline-proline bond
in the cyclin E phosphodegron (P382) (56). Then, being mediated by nucleolar
protein nucleophosmin (NPM), the complex binds to
SCFFbxw7γ and translocates from the nucleoplasm into the
nucleolus where cyclin E is multiubiquitylated, but does not
execute proteasomal degradation (87). Notably, Cdk2 kinase was recently
demonstrated to interact with Fbxw7γ, result in Fbxw7γ degradation
(89). This system serves a novel
mechanism for rapid inactivation of cyclin E, through separation of
cyclin E from its targets.
Some studies have focused on the upstream of the
Fbxw7/cyclin E pathway. Their findings have identified many factors
that increase or decrease Fbxw7 expression, thus regulating the
Fbxw7/cyclin E pathway (Fig. 1).
The PI3K/Akt pathway, which was previously thought to be pivotal
for the regulation of the GSK phosphorylation, was recently
reported to mediate the phosphorylation of Fbxw7α at S227 (90). This modification stabilizes Fbxw7
and promotes ubiquitylation of the two substrates, cyclin E and
c-Myc (90). In addition, Sim et
al reported a novel mechanism whereby TRIP-Br proteins links to
E2F to act as upstream regulators of the Fbxw7/cyclin E pathway in
the maintenance of genomic stability (91). This mechanism reveals a function
distinct from the conventional function of E2F, which increases
cyclin E expression at the transcriptional level. TRIP-Br proteins
interact with PHD zinc finger and/or bromodomain proteins such as
KRIP-1 and p300/CBP, upregulate the FBXW7 gene product via
the activation of E2F transcriptional activity, and subsequently
lead to downregulation of the cyclin E protein (91). Polo-like kinase 2 (PIK2) is also
found to directly phosphorylate Fbxw7 at S25, S176 and S349.
However, these phosphorylations reduce the stability of Fbxw7, thus
stabilizing cyclin E, and contributing to the duplication of
centrioles and aneuploidy (92).
TAL1, a class II basic helix-loop-helix (bHLH) transcription
factor, promotes the malignant phenotype in T-ALL through the
repression of Fbxw7 in a miR223-dependent manner. This effect leads
to a marked increase of the expression of cyclin E and c-Myc
(93).
The most distinctive function of cyclin E is to bind
to and activate Cdk2. Activation of the cyclin E/Cdk2 complex
subsequently leads to the phosphorylation of its substrates, such
as retinoblastoma (81), cdc6, NPM,
p21 and p27 (72). Of note, cyclin
E/Cdk2 was reported to directly phosphorylate Cdh1, thus
inactivating APCCdh1, an E3 ligase important for genomic
stability (94). Furthermore, Lau
et al showed that the Fbxw7/cyclin E pathway regulates the
activation of APCCdh1 through direct phosphorylation of
Cdh1 (95). This subsequently leads
to the inactivation of APCCdh1 E3 ligase and
upregulation of APCCdh1-specific substrates, which are
well-characterized oncoproteins (Fig.
1). These results suggested that Fbxw7 regulates cell cycle
through, not only cyclin E itself, but also the downstream
substrates of cyclin E/Cdk2 complex.
Taken together, these findings suggest that
Fbxw7/cyclin E is regulated by multiple regulators, which partly
explains why there are no FBXW7 gene mutations in some
tumors. Moreover, these findings provide insight into the
tumor-suppressive function of Fbxw7.
The c-Myc protein, a basic helix-loop-helix zipper
(bHLH/Zip)-type transcription factor, generally combines with its
cofactor MAX and activates their target genes transcription by
binding E-box motifs (CACGTG), and thus playing a predominant role
in cell proliferation and tumorigenesis (96). Accumulated evidence has indicated
that c-Myc protein turnover is tightly regulated at the
post-translational level through ubiquitin-proteasome pathway
controlled by the SCFFbxw7 complex (97,98).
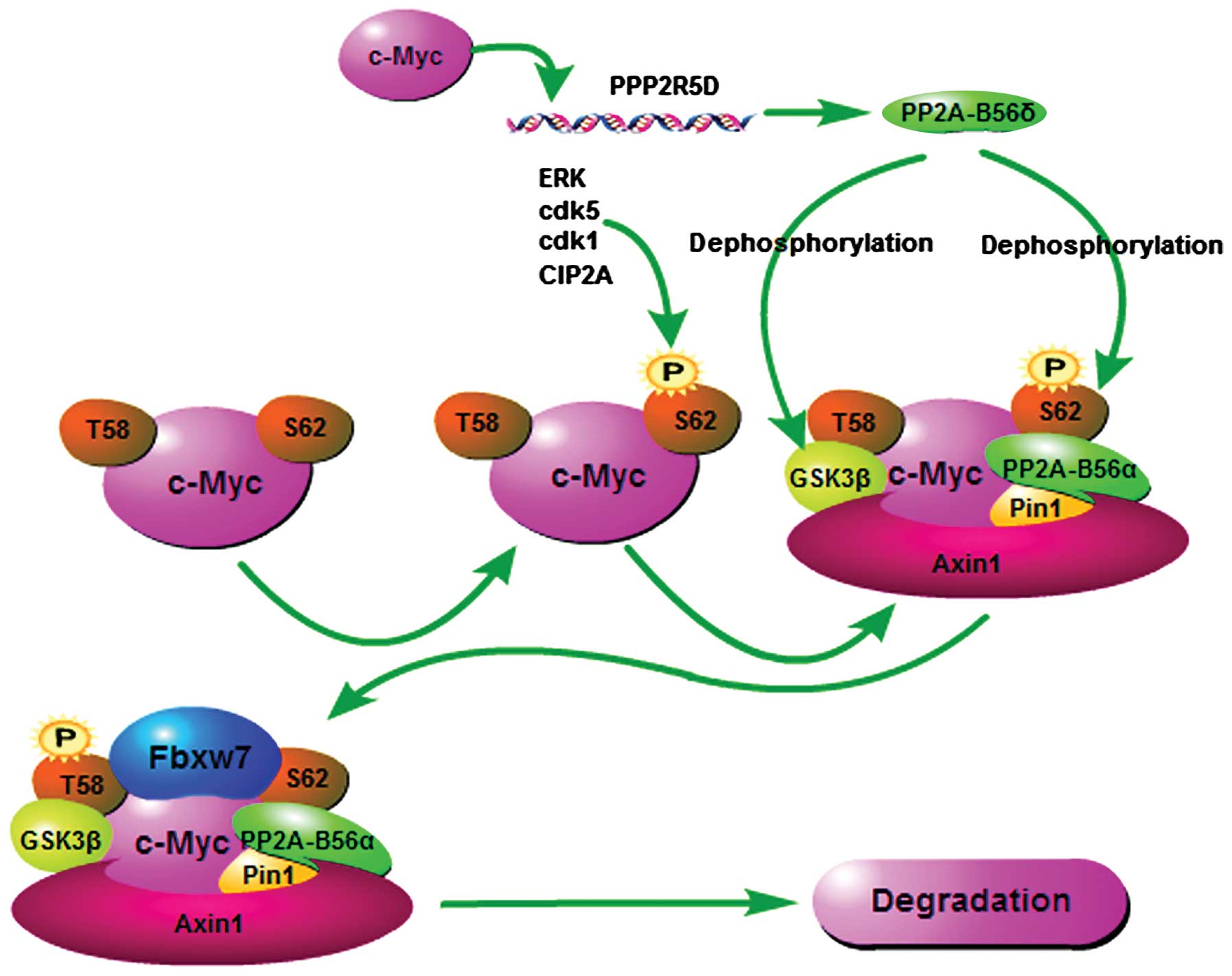
The oncoprotein c-Myc has only one CPD, containing the sequence
PTPPLSP (residues 57–63 in human c-Myc), within which T58 and S62
are the phosphorylation sites (98). However, these two phosphorylation
sites exert opposite functions on c-Myc degradation, as the
phosphorylation of S62 results in c-Myc stabilization, whereas the
T58 phosphorylation by GSK3β contributes to the interaction between
Fbxw7 and c-Myc (98,99). The phosphorylation of T58 depends on
the prime phosphorylation of Ser-62. This phosphorylation-dependent
proteolysis controlled by SCFFbxw7 is involved with
complicated feedback mechanisms.
The activation of Ras-dependent phosphorylation
pathways is considered to be important for c-Myc stability mainly
through two effective pathways: the Raf/MEK/ERK pathway stabilizes
c-Myc by enhancing S62 phosphorylation, and the PI3K/Akt pathway
which disrupts the interaction between Fbxw7 and c-Myc by
phosphorylating GSK3β at S9, which in turn decreases the
phosphorylation of T58 within c-Myc (99). Thus in quiescent cells, where the
growth stimulus disappears, Ras activity declines, and the activity
of the PI3K/Akt pathway is also downregulated, resulting in the
enhancement of T58 phosphorylation and the degradation of c-Myc. It
has been shown that the dephosphorylation of S62 is also crucial
for the ubiquitin-mediated degradation of c-Myc. T58
phosphorylation is found to facilitate the interaction between
c-Myc and Pin1 which catalyzes the isomerization of proline
residues in c-Myc to promote S62 dephosphorylation by protein
phosphatase 2A (PP2A) (100).
Further study showed that Axin1, a multi-domain scaffold protein,
regulates this process by facilitating the association of GSK3β,
B56α (one of the regulatory subunits of PP2A), and Pin1 with c-Myc,
forming an Axin-Pin1-GSK3β-PP2A/B56α complex to promote the
ubiquitin-mediated degradation of c-Myc (101). c-Myc has been reported to bind to
and transcriptionally activate the PPP2R5D gene that encodes
B56δ (another regulatory subunit of PP2A) (102). PP2A-B56δ, not only S
dephosphorylates S62 within c-Myc in the same manner as PP2A-B56α,
but also reverses the GSK3β inhibitory phosphorylation at S9 by
PI3K/Akt (102). These studies
provide a mechanism that links c-Myc protein degradation controlled
by Fbxw7 to a complicated feedback pathway. In addition, Cdk5, Cdk1
and CIP2A increase S62 of c-Myc phosphorylation in a direct or
indirect manner, resulting in c-Myc stabilization (103–105) (Fig.
3).
Similar to cyclin E, the Fbxw7/c-Myc pathway appears
to be linked to many signal molecules. Previous findings have shown
that rictor binds to Fbxw7 and facilitates ubiquitination of c-Myc
(78). In addition to rictor, other
interaction partners such as stomatin-like protein 1 (SLP-1), NPM,
and Bloom (BLM) forms complexes with Fbxw7; in particular, Fbxw7γ,
which is specific for the ubiquitination of c-Myc (89,106–108). By performing a two-hybrid screen,
Zhang et al identified that SLP-1, as a novel interaction
partner, can bind to the N-terminus of Fbxw7γ and stabilize Fbxw7γ,
leading to an even greater reduction in c-Myc abundance (89). NPM is required by Fbxw7γ for the
proper folding, nucleolar localization and stabilization. Mutation
of NPM induces delocalization and destabilization of Fbxw7γ and
stabilization of c-Myc (106).
Recently, BLM, which is a helicase mutated in Bloom syndrome and is
conclusively regarded as a sensor of DNA lesion, was reported to
have contact with c-Myc and Fbxw7, leading to the degradation of
c-Myc and subsequent delay of colorectal tumorigenesis (108,109).
By contrast, many factors serve as binding partners
of c-Myc, and block the degradation of c-Myc in a Fbxw7-dependent
manner. For example, NF-κB essential modulator (NEMO) induces the
upregulation of c-Myc protein through direct interaction with c-Myc
protein and inhibits ubiquitination activity of Fbxw7 without
interfering with the interaction of Fbxw7 with T58-phoshorylated
c-Myc (110).
Although some studies focus on the interaction
mechanism between Fbxw7 and c-Myc, other studies provide
mechanistic insights for the regulation of the Fbxw7/c-Myc pathway
through a reduction of Fbxw7 expression. For example, NF-κB1 (p50),
a ubiquitously expressed subunit of NF-κB, is thought to suppress
FBXW7 gene transcription and upregulate c-Myc protein
expression (111). Huang et
al revealed that Fbxw7 was profoundly upregulated in
p50-deficient cells in comparison to that in p50 intact cells,
whereas knockdown of Fbxw7 in p50−/− cells restored
arsenite-induced c-Myc protein accumulation (111). COP9 signalosome (CSN) has been
found to facilitate the autoubiquitination/degradation of Fbxw7,
thereby stabilizing c-Myc (112).
Similar to protein factors, some miRNAs participate in the
regulation of c-Myc by interfering with the expression of Fbxw7.
For example, miR-92 mediates the proteolytic degradation of c-Myc
by direct repression of Fbxw7 in a Eµ-myc Burkitt's lymphoma
model (113). Thus, miR-92
overexpression leads to increase of aberrant c-Myc.
Taken together, the abovementioned studies showed
that the Fbxw7/c-Myc pathway is regulated by accurate mechanisms.
These mechanisms ensure the cell cycle progression in normal cells
and regulate cell proliferation and tumorigenesis in malignant
tumors. Moreover, these regulation mechanisms (regulation of the
Fbxw7/cylin E and Fbxw7/c-Myc pathways) may also exist in other
Fbxw7-specific substrates. Furthermore, the fact that some
regulators are involved in the Fbxw7/cyclin E and Fbxw7/c-Myc
pathways suggests that the two pathways are important for tumor
cell proliferation, and the abnormality of these pathways may be
partially regulated by the same mechanisms.
As a tumor suppressor, mRNA and protein expression
levels of Fbxw7 have been shown to be downregulated in various
types of human cancer. Fbxw7 has been found to be inactivated by
mutation in several malignancies with an overall mutation frequency
of ~6% (18). However, no studies
have yet reported Fbxw7 mutations in HCC, one of the leading causes
of cancer-related mortality worldwide. Notably, Fbxw7 expression is
reported to be important in the regulation of lipogenesis and cell
proliferation and differentiation in the liver (114). Using liver-specific Fbxw7 null
mice, Onoyama et al found that the hepatic ablation of Fbxw7
resulted in hepatomegaly and steatohepatitis, and long-term Fbxw7
deficiency resulted in marked proliferation of the biliary system
and development of hamartomas (114). Moreover, double heterozygous
p53−/− Fbxw7−/− mice have been proven to
develop hepatocarcinomas (35).
These results suggest that Fbxw7 is critical for the liver
development and tumorigenesis.
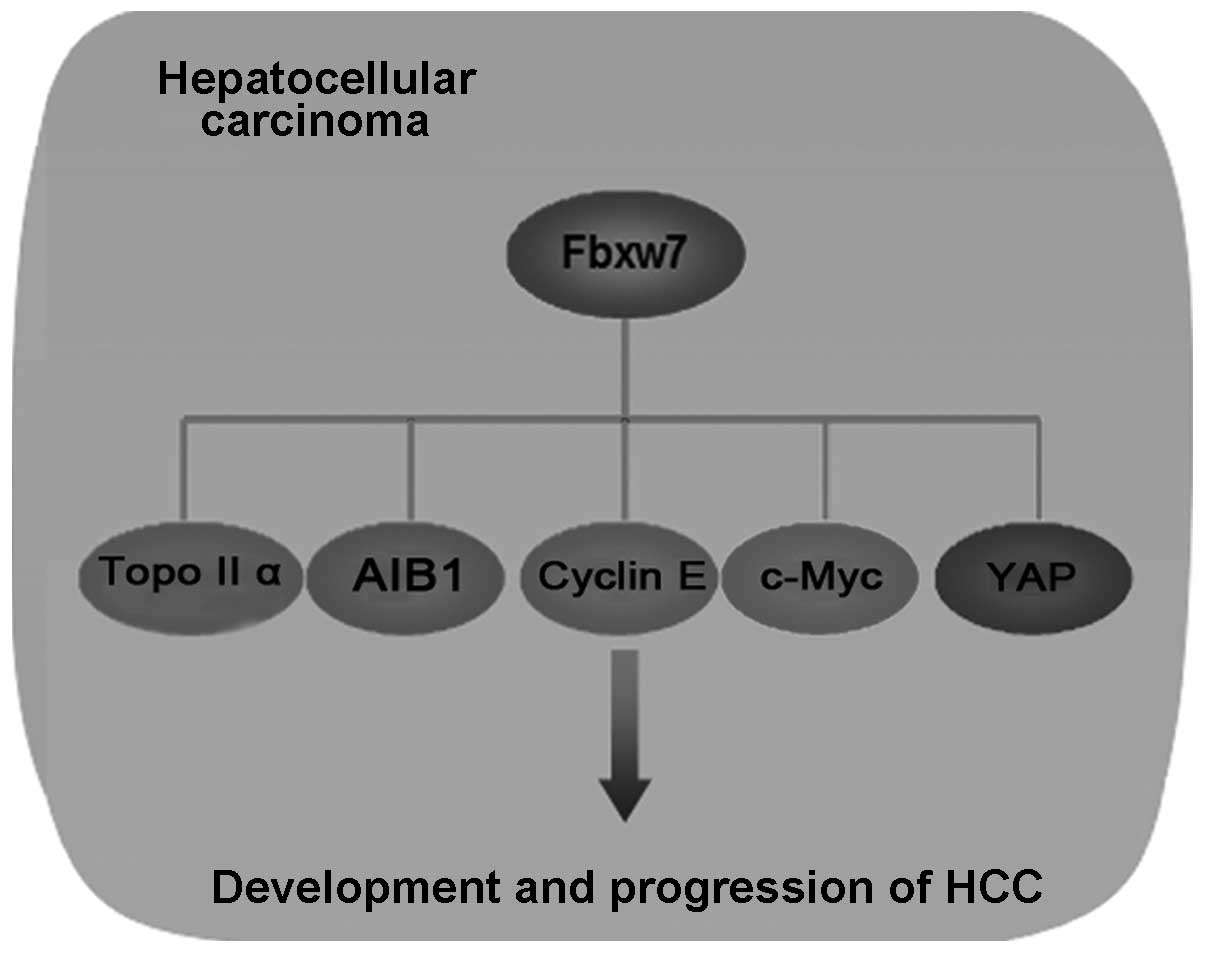
Previous studies from different groups have
identified the role of Fbxw7 as a tumor suppressor in HCC. Chen
et al were the first group to investigate Fbxw7 in HCC
(47). Their findings revealed that
Fbxw7 promoted ubiquitin-dependent degradation of TopoIIα in a
HADC-dependent manner (47). Liu
et al found that Fbxw7 was involved in the
ubiquitin-dependent degradation of AIB1 in Hbx-related HCC
(31). Notably, Tu et al
were the first group to report that the Fbxw7 mRNA and protein
expression in HCC tissues was significantly lower than that in
normal tumor-adjacent tissues (115). Subsequently, they also identified
that c-Myc, cyclin E, and YAP proteins abundance in HCC was
regulated by Fbxw7 (53,116). Consistent with Tu et al,
Imura et al reported a lower Fbxw7 expression in HCC tissues
compared with non-tumor liver tissues (117). Coincidentally, all these
substrates of Fbxw7 in HCC are closely associated with promoting
cancer cell proliferation and tumorigenesis (53,71,96,118,119). Given that c-Myc, YAP and AIB1 have
transcriptional activity and promote the transcription of many
oncogenes with different functions, Fbxw7 may be associated with
controlling cell growth and regulating other malignant behavior,
such as invasion and metastasis (96,120,121) (Fig.
4). Taken together, these results show that Fbxw7 plays a
critical role in HCC. The inactivation of Fbxw7 in HCC is involved
in tumorigenesis. However, in-depth investigation is required to
determine how Fbxw7 is inactivated in HCC and whether other signal
pathways, through which Fbxw7 plays as a tumor suppressor in HCC,
exist.
Most cell proteins involved in cell cycle
progression, proliferation and apoptosis are regulated by the UPS,
which consist of three classes of enzymes (E1, E2 and E3). The
SCFFbxw7 complex is one of the most well-known E3
ligases. Fbxw7, as the substrates-recognition component of the
SCFFbxw7 complex, regulates cell proliferation, genetic
stability, and tumorigenesis in humans by coordinating the
ubiquitin-dependent proteolysis of several key oncoproteins.
Several studies (refs?) have identified a growing list of specific
substrates of Fbxw7, such as YAP and Eya1. However, cyclin E and
c-Myc are the best characterized oncoproteins among the substrates
of Fbxw7. Multiple factors tightly regulate the Fbxw7/cyclin E and
Fbxw7/c-Myc pathways in different mechanisms. These regulation
mechanisms may also exist in other Fbxw7-specific substrates. These
mechanisms therefore may be useful in understanding the functions
of Fbxw7-related signaling pathways in the regulation of cell
proliferation and tumorigenesis in cancer. Furthermore, this
understanding reveals Fbxw7-related signaling pathways have
potential in developing new targets in cancer therapy. In addition,
since many studies have reported that Fbxw7 functions as a tumor
suppressor and plays a critical role in regulating several key
oncoproteins in HCC, Fbxw7 is a potential therapeutic target in
this cancer. However, future studies should be conducted to
invesigate the mechanism of Fbxw7 and how it may serve as a tumor
suppressor in HCC.
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81301768 and
81372565). The authors would like to thank Professor Qingguang Liu
and Dr Kangsheng Tu (Xi'an Jiaotong University, Xi'an, China) for
their technical assistance.
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al CONCORD Working Group: Global surveillance of cancer survival
1995–2009: Analysis of individual data for 25,676,887 patients from
279 population-based registries in 67 countries (CONCORD-2).
Lancet. 385:977–1010. 2015. View Article : Google Scholar
|
|
3
|
Adams J: Development of the proteasome
inhibitor PS-341. Oncologist. 7:9–16. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tu Y, Chen C, Pan J, Xu J, Zhou ZG and
Wang CY: The Ubiquitin Proteasome Pathway (UPP) in the regulation
of cell cycle control and DNA damage repair and its implication in
tumorigenesis. Int J Clin Exp Pathol. 5:726–738. 2012.PubMed/NCBI
|
|
5
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011. View
Article : Google Scholar
|
|
6
|
Voutsadakis IA: The ubiquitin-proteasome
system and signal transduction pathways regulating Epithelial
Mesenchymal transition of cancer. J Biomed Sci. 19:672012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Devoy A, Soane T, Welchman R and Mayer RJ:
The ubiquitin-proteasome system and cancer. Essays Biochem.
41:187–203. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Komander D: The emerging complexity of
protein ubiquitination. Biochem Soc Trans. 37:937–953. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pickart CM: Mechanisms underlying
ubiquitination. Annu Rev Biochem. 70:503–533. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng N, Schulman BA, Song L, Miller JJ,
Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al:
Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase
complex. Nature. 416:703–709. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schulman BA, Carrano AC, Jeffrey PD, Bowen
Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M and
Pavletich NP: Insights into SCF ubiquitin ligases from the
structure of the Skp1-Skp2 complex. Nature. 408:381–386. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hartwell LH, Mortimer RK, Culotti J and
Culotti M: Genetic control of the cell division cycle in yeast: V.
genetic analysis of cdc mutants. Genetics. 74:267–286.
1973.PubMed/NCBI
|
|
13
|
Hubbard EJ, Wu G, Kitajewski J and
Greenwald I: sel-10, a negative regulator of lin-12 activity in
Caenorhabditis elegans, encodes a member of the CDC4 family of
proteins. Genes Dev. 11:3182–3193. 1997. View Article : Google Scholar
|
|
14
|
Moberg KH, Bell DW, Wahrer DC, Haber DA
and Hariharan IK: Archipelago regulates Cyclin E levels in
Drosophila and is mutated in human cancer cell lines. Nature.
413:311–316. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Welcker M and Clurman BE: FBW7 ubiquitin
ligase: A tumour suppressor at the crossroads of cell division,
growth and differentiation. Nat Rev Cancer. 8:83–93. 2008.
View Article : Google Scholar
|
|
16
|
Rajagopalan H, Jallepalli PV, Rago C,
Velculescu VE, Kinzler KW, Vogelstein B and Lengauer C:
Inactivation of hCDC4 can cause chromosomal instability. Nature.
428:77–81. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Spruck CH, Strohmaier H, Sangfelt O,
Müller HM, Hubalek M, Müller-Holzner E, Marth C, Widschwendter M
and Reed SI: hCDC4 gene mutations in endometrial cancer. Cancer
Res. 62:4535–4539. 2002.PubMed/NCBI
|
|
18
|
Akhoondi S, Sun D, von der Lehr N,
Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D,
Marth C, et al: FBXW7/hCDC4 is a general tumor suppressor in human
cancer. Cancer Res. 67:9006–9012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davis H, Lewis A, Behrens A and Tomlinson
I: Investigation of the atypical FBXW7 mutation spectrum in human
tumours by conditional expression of a heterozygous propellor tip
missense allele in the mouse intestines. Gut. 63:792–799. 2014.
View Article : Google Scholar :
|
|
20
|
Koepp DM, Schaefer LK, Ye X, Keyomarsi K,
Chu C, Harper JW and Elledge SJ: Phosphorylation-dependent
ubiquitination of cyclin E by the SCFFbw7 ubiquitin
ligase. Science. 294:173–177. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yada M, Hatakeyama S, Kamura T, Nishiyama
M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K and
Nakayama KI: Phosphorylation-dependent degradation of c-Myc is
mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei W, Jin J, Schlisio S, Harper JW and
Kaelin WG Jr: The v-Jun point mutation allows c-Jun to escape
GSK3-dependent recognition and destruction by the Fbw7 ubiquitin
ligase. Cancer Cell. 8:25–33. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
O'Neil J, Grim J, Strack P, Rao S,
Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters
R, et al: FBW7 mutations in leukemic cells mediate NOTCH pathway
activation and resistance to gamma-secretase inhibitors. J Exp Med.
204:1813–1824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCFFBW7 regulates cellular apoptosis by targeting MCL1
for ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mao JH, Kim IJ, Wu D, Climent J, Kang HC,
DelRosario R and Balmain A: FBXW7 targets mTOR for degradation and
cooperates with PTEN in tumor suppression. Science. 321:1499–1502.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Pauley AM, Myers RL, Shuang R,
Brashler JR, Yan R, Buhl AE, Ruble C and Gurney ME: SEL-10
interacts with presenilin 1, facilitates its ubiquitination, and
alters A-beta peptide production. J Neurochem. 82:1540–1548. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brockmann M, Poon E, Berry T, Carstensen
A, Deubzer HE, Rycak L, Jamin Y, Thway K, Robinson SP, Roels F, et
al: Small molecule inhibitors of aurora-a induce proteasomal
degradation of N-myc in childhood neuroblastoma. Cancer Cell.
24:75–89. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Galli F, Rossi M, D'Alessandra Y, De
Simone M, Lopardo T, Haupt Y, Alsheich-Bartok O, Anzi S, Shaulian
E, Calabrò V, et al: MDM2 and Fbw7 cooperate to induce p63 protein
degradation following DNA damage and cell differentiation. J Cell
Sci. 123:2423–2433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kitagawa K, Hiramatsu Y, Uchida C, Isobe
T, Hattori T, Oda T, Shibata K, Nakamura S, Kikuchi A and Kitagawa
M: Fbw7 promotes ubiquitin-dependent degradation of c-Myb:
Involvement of GSK3-mediated phosphorylation of Thr-572 in mouse
c-Myb. Oncogene. 28:2393–2405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cassavaugh JM, Hale SA, Wellman TL, Howe
AK, Wong C and Lounsbury KM: Negative regulation of HIF-1α by an
FBW7-mediated degradation pathway during hypoxia. J Cell Biochem.
112:3882–3890. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y, Tong Z, Li T, Chen Q, Zhuo L, Li W,
Wu RC and Yu C: Hepatitis B virus X protein stabilizes amplified in
breast cancer 1 protein and cooperates with it to promote human
hepatocellular carcinoma cell invasiveness. Hepatology.
56:1015–1024. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tu K, Zheng X, Yin G, Zan X, Yao Y and Liu
Q: Evaluation of Fbxw7 expression and its correlation with
expression of SREBP-1 in a mouse model of NAFLD. Mol Med Rep.
6:525–530. 2012.PubMed/NCBI
|
|
33
|
Pérez-Benavente B, García JL, Rodríguez
MS, Pineda-Lucena A, Piechaczyk M, Font de Mora J and Farràs R:
GSK3-SCFFBXW7 targets JunB for degradation in G2 to
preserve chromatid cohesion before anaphase. Oncogene.
32:2189–2199. 2013. View Article : Google Scholar
|
|
34
|
Arabi A, Ullah K, Branca RM, Johansson J,
Bandarra D, Haneklaus M, Fu J, Ariës I, Nilsson P, Den Boer ML, et
al: Proteomic screen reveals Fbw7 as a modulator of the NF-κB
pathway. Nat Commun. 3:9762012. View Article : Google Scholar
|
|
35
|
Mao JH, Perez-Losada J, Wu D, Delrosario
R, Tsunematsu R, Nakayama KI, Brown K, Bryson S and Balmain A:
Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor
gene. Nature. 432:775–779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Teng CL, Hsieh YC, Phan L, Shin J, Gully
C, Velazquez-Torres G, Skerl S, Yeung SC, Hsu SL and Lee MH: FBXW7
is involved in Aurora B degradation. Cell Cycle. 11:4059–4068.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao J, Tang J, Men W and Ren K:
FBXW7-mediated degradation of CCDC6 is impaired by ATM during DNA
damage response in lung cancer cells. FEBS Lett. 586:4257–4263.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Davis MA, Larimore EA, Fissel BM, Swanger
J, Taatjes DJ and Clurman BE: The SCF-Fbw7 ubiquitin ligase
degrades MED13 and MED13L and regulates CDK8 module association
with Mediator. Genes Dev. 27:151–156. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan M, Zhao Y, Kim SJ, Liu M, Jia L,
Saunders TL, Zhu Y and Sun Y: SAG/RBX2/ROC2 E3 ubiquitin ligase is
essential for vascular and neural development by targeting NF1 for
degradation. Dev Cell. 21:1062–1076. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang R, Wang Y, Liu N, Ren C, Jiang C,
Zhang K, Yu S, Chen Y, Tang H, Deng Q, et al: FBW7 regulates
endothelial functions by targeting KLF2 for ubiquitination and
degradation. Cell Res. 23:803–819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bialkowska AB, Liu Y, Nandan MO and Yang
VW: A colon cancer-derived mutant of Krüppel-like factor 5 (KLF5)
is resistant to degradation by glycogen synthase kinase 3β (GSK3β)
and the E3 ubiquitin ligase F-box and WD repeat domain-containing
7α (FBW7α). J Biol Chem. 289:5997–6005. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bengoechea-Alonso MT and Ericsson J: The
ubiquitin ligase Fbxw7 controls adipocyte differentiation by
targeting C/EBPalpha for degradation. Proc Natl Acad Sci USA.
107:11817–11822. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Balamurugan K, Sharan S, Klarmann KD,
Zhang Y, Coppola V, Summers GH, Roger T, Morrison DK, Keller JR and
Sterneck E: FBXW7α attenuates inflammatory signalling by
downregulating C/EBPδ and its target gene Tlr4. Nat Commun.
4:16622013. View Article : Google Scholar
|
|
44
|
Biswas M, Phan D, Watanabe M and Chan JY:
The Fbw7 tumor suppressor regulates nuclear factor E2-related
factor 1 transcription factor turnover through proteasome-mediated
proteolysis. J Biol Chem. 286:39282–39289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lochab S, Pal P, Kapoor I, Kanaujiya JK,
Sanyal S, Behre G and Trivedi AK: E3 ubiquitin ligase Fbw7
negatively regulates granulocytic differentiation by targeting
G-CSFR for degradation. Biochim Biophys Acta. 1833:2639–2652. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yumimoto K, Matsumoto M, Onoyama I,
Imaizumi K and Nakayama KI: F-box and WD repeat domain-containing-7
(Fbxw7) protein targets endoplasmic reticulum-anchored osteogenic
and chondrogenic transcriptional factors for degradation. J Biol
Chem. 288:28488–28502. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen MC, Chen CH, Chuang HC, Kulp SK, Teng
CM and Chen CS: Novel mechanism by which histone deacetylase
inhibitors facilitate topoisomerase IIα degradation in
hepatocellular carcinoma cells. Hepatology. 53:148–159. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bengoechea-Alonso MT and Ericsson J: Tumor
suppressor Fbxw7 regulates TGFβ signaling by targeting TGIF1 for
degradation. Oncogene. 29:5322–5328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun Y and Li X: The canonical wnt signal
restricts the glycogen synthase kinase 3/fbw7-dependent
ubiquitination and degradation of eya1 phosphatase. Mol Cell Biol.
34:2409–2417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kitagawa K, Shibata K, Matsumoto A,
Matsumoto M, Ohhata T, Nakayama KI, Niida H and Kitagawa M: Fbw7
targets GATA3 through cyclin-dependent kinase 2-dependent
proteolysis and contributes to regulation of T-cell development.
Mol Cell Biol. 34:2732–2744. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dai X, North BJ and Inuzuka H: Negative
regulation of DAB2IP by Akt and SCFFbw7 pathways.
Oncotarget. 5:3307–3315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Trausch-Azar JS, Abed M, Orian A and
Schwartz AL: Isoform-specific SCFFbw7 ubiquitination
mediates differential regulation of PGC-1α. J Cell Physiol.
230:842–852. 2015. View Article : Google Scholar
|
|
53
|
Tu K, Yang W, Li C, Zheng X, Lu Z, Guo C,
Yao Y and Q: Fbxw7 is an independent prognostic marker and induces
apoptosis and growth arrest by regulating YAP abundance in
hepatocellular carcinoma. Mol Cancer. 13:1102014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jin J, Cardozo T, Lovering RC, Elledge SJ,
Pagano M and Harper JW: Systematic analysis and nomenclature of
mammalian F-box proteins. Genes Dev. 18:2573–2580. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sterian A, Kan T, Berki AT, Mori Y, Olaru
A, Schulmann K, Sato F, Wang S, Paun B, Cai K, et al: Mutational
and LOH analyses of the chromosome 4q region in esophageal
adenocarcinoma. Oncology. 70:168–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
van Drogen F, Sangfelt O, Malyukova A,
Matskova L, Yeh E, Means AR and Reed SI: Ubiquitylation of cyclin E
requires the sequential function of SCF complexes containing
distinct hCdc4 isoforms. Mol Cell. 23:37–48. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Matsumoto A, Tateishi Y, Onoyama I, Okita
Y, Nakayama K and Nakayama KI: Fbxw7β resides in the endoplasmic
reticulum membrane and protects cells from oxidative stress. Cancer
Sci. 102:749–755. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ren H, Zhao L, Li Y, Yue P, Deng X,
Owonikoko TK, Chen M, Khuri FR and Sun SY: The PI3 kinase inhibitor
NVP-BKM120 induces GSK3/FBXW7-dependent Mcl-1 degradation,
contributing to induction of apoptosis and enhancement of
TRAIL-induced apoptosis. Cancer Lett. 338:229–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Embi N, Rylatt DB and Cohen P: Glycogen
synthase kinase-3 from rabbit skeletal muscle. Separation from
cyclic-AMP-dependent protein kinase and phosphorylase kinase. Eur J
Biochem. 107:519–527. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Woodgett JR: Molecular cloning and
expression of glycogen synthase kinase-3/factor A. EMBO J.
9:2431–2438. 1990.PubMed/NCBI
|
|
61
|
Wu D and Pan W: GSK3: A multifaceted
kinase in Wnt signaling. Trends Biochem Sci. 35:161–168. 2010.
View Article : Google Scholar :
|
|
62
|
Kim L and Kimmel AR: GSK3, a master switch
regulating cell-fate specification and tumorigenesis. Curr Opin
Genet Dev. 10:508–514. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Buttrick GJ and Wakefield JG: PI3-K and
GSK-3: Akt-ing together with microtubules. Cell Cycle. 7:2621–2625.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Welcker M, Singer J, Loeb KR, Grim J,
Bloecher A, Gurien-West M, Clurman BE and Roberts JM: Multisite
phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol
Cell. 12:381–392. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hao B, Oehlmann S, Sowa ME, Harper JW and
Pavletich NP: Structure of a Fbw7-Skp1-cyclin E complex:
Multisite-phosphorylated substrate recognition by SCF ubiquitin
ligases. Mol Cell. 26:131–143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bahram F, von der Lehr N, Cetinkaya C and
Larsson LG: c-Myc hot spot mutations in lymphomas result in
inefficient ubiquitination and decreased proteasome-mediated
turnover. Blood. 95:2104–2110. 2000.PubMed/NCBI
|
|
67
|
Tan Y, Sangfelt O and Spruck C: The
Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett.
271:1–12. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Strohmaier H, Spruck CH, Kaiser P, Won KA,
Sangfelt O and Reed SI: Human F-box protein hCdc4 targets cyclin E
for proteolysis and is mutated in a breast cancer cell line.
Nature. 413:316–322. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ye X, Nalepa G, Welcker M, Kessler BM,
Spooner E, Qin J, Elledge SJ, Clurman BE and Harper JW: Recognition
of phosphodegron motifs in human cyclin E by the SCFFbw7
ubiquitin ligase. J Biol Chem. 279:50110–50119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Welcker M and Clurman BE: Fbw7/hCDC4
dimerization regulates its substrate interactions. Cell Div.
2:72007. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Geng Y, Lee YM, Welcker M, Swanger J,
Zagozdzon A, Winer JD, Roberts JM, Kaldis P, Clurman BE and
Sicinski P: Kinase-independent function of cyclin E. Mol Cell.
25:127–139. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Siu KT, Rosner MR and Minella AC: An
integrated view of cyclin E function and regulation. Cell Cycle.
11:57–64. 2012. View Article : Google Scholar :
|
|
73
|
Minella AC, Grim JE, Welcker M and Clurman
BE: p53 and SCFFbw7 cooperatively restrain cyclin
E-associated genome instability. Oncogene. 26:6948–6953. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Minella AC, Swanger J, Bryant E, Welcker
M, Hwang H and Clurman BE: p53 and p21 form an inducible barrier
that protects cells against cyclin E-cdk2 deregulation. Curr Biol.
12:1817–1827. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kimura T, Gotoh M, Nakamura Y and Arakawa
H: hCDC4b, a regulator of cyclin E, as a direct transcriptional
target of p53. Cancer Sci. 94:431–436. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mandal S, Freije WA, Guptan P and Banerjee
U: Metabolic control of G1-S transition: Cyclin E degradation by
p53-induced activation of the ubiquitin-proteasome system. J Cell
Biol. 188:473–479. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Finkin S, Aylon Y, Anzi S, Oren M and
Shaulian E: Fbw7 regulates the activity of endoreduplication
mediators and the p53 pathway to prevent drug-induced polyploidy.
Oncogene. 27:4411–4421. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Guo Z, Zhou Y, Evers BM and Wang Q: Rictor
regulates FBXW7-dependent c-Myc and cyclin E degradation in
colorectal cancer cells. Biochem Biophys Res Commun. 418:426–432.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang H, Zhang X, Geng L, Teng L and
Legerski RJ: Artemis regulates cell cycle recovery from the S phase
checkpoint by promoting degradation of cyclin E. J Biol Chem.
284:18236–18243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Welcker M and Clurman BE: The SV40 large T
antigen contains a decoy phosphodegron that mediates its
interactions with Fbw7/hCdc4. J Biol Chem. 280:7654–7658. 2005.
View Article : Google Scholar
|
|
81
|
Sarbassov DD, Ali SM, Kim DH, Guertin DA,
Latek RR, Erdjument-Bromage H, Tempst P and Sabatini DM: Rictor, a
novel binding partner of mTOR, defines a rapamycin-insensitive and
raptor-independent pathway that regulates the cytoskeleton. Curr
Biol. 14:1296–1302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Poinsignon C, de Chasseval R, Soubeyrand
S, Moshous D, Fischer A, Haché RJ and de Villartay JP:
Phosphorylation of Artemis following irradiation-induced DNA
damage. Eur J Immunol. 34:3146–3155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ahuja D, Sáenz-Robles MT and Pipas JM:
SV40 large T antigen targets multiple cellular pathways to elicit
cellular transformation. Oncogene. 24:7729–7745. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Minella AC, Welcker M and Clurman BE: Ras
activity regulates cyclin E degradation by the Fbw7 pathway. Proc
Natl Acad Sci USA. 102:9649–9654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hynes NE and Lane HA: ERBB receptors and
cancer: The complexity of targeted inhibitors. Nat Rev Cancer.
5:341–354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Tan Y, Sun D, Jiang W, Klotz-Noack K,
Vashisht AA, Wohlschlegel J, Widschwendter M and Spruck C:
PP2A-B55β antagonizes cyclin E1 proteolysis and promotes its
dysregulation in cancer. Cancer Res. 74:2006–2014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bhaskaran N, van Drogen F, Ng HF, Kumar R,
Ekholm-Reed S, Peter M, Sangfelt O and Reed SI: Fbw7α and Fbw7γ
collaborate to shuttle cyclin E1 into the nucleolus for
multiubiquitylation. Mol Cell Biol. 33:85–97. 2013. View Article : Google Scholar :
|
|
88
|
Reed SI: Cooperation between different
Cdc4/Fbw7 isoforms may be associated with 2-step inactivation of
SCFCdc4 targets. Cell Cycle. 5:1923–1924. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhang W, MacDonald EM and Koepp DM: The
stomatin-like protein SLP-1 and Cdk2 interact with the F-Box
protein Fbw7-γ. PLoS One. 7:e477362012. View Article : Google Scholar
|
|
90
|
Schülein C, Eilers M and Popov N:
PI3K-dependent phosphorylation of Fbw7 modulates substrate
degradation and activity. FEBS Lett. 585:2151–2157. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Sim KG, Zang Z, Yang CM, Bonventre JV and
Hsu SI: TRIP-Br links E2F to novel functions in the regulation of
cyclin E expression during cell cycle progression and in the
maintenance of genomic stability. Cell Cycle. 3:1296–1304. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Cizmecioglu O, Krause A, Bahtz R, Ehret L,
Malek N and Hoffmann I: Plk2 regulates centriole duplication
through phosphorylation-mediated degradation of Fbxw7 (human Cdc4).
J Cell Sci. 125:981–992. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Mansour MR, Sanda T, Lawton LN, Li X,
Kreslavsky T, Novina CD, Brand M, Gutierrez A, Kelliher MA,
Jamieson CH, et al: The TAL1 complex targets the FBXW7 tumor
suppressor by activating miR-223 in human T cell acute
lymphoblastic leukemia. J Exp Med. 210:1545–1557. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Keck JM, Summers MK, Tedesco D,
Ekholm-Reed S, Chuang LC, Jackson PK and Reed SI: Cyclin E
overexpression impairs progression through mitosis by inhibiting
APCCdh1. J Cell Biol. 178:371–385. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lau AW, Inuzuka H, Fukushima H, Wan L, Liu
P, Gao D, Sun Y and Wei W: Regulation of APCCdh1 E3
ligase activity by the Fbw7/cyclin E signaling axis contributes to
the tumor suppressor function of Fbw7. Cell Res. 23:947–961. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Meyer N and Penn LZ: Reflecting on 25
years with MyC. Nat Rev Cancer. 8:976–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Onoyama I, Tsunematsu R, Matsumoto A,
Kimura T, de Alborán IM, Nakayama K and Nakayama KI: Conditional
inactivation of Fbxw7 impairs cell-cycle exit during T cell
differentiation and results in lymphomatogenesis. J Exp Med.
204:2875–2888. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Welcker M, Orian A, Jin J, Grim JE, Harper
JW, Eisenman RN and Clurman BE: The Fbw7 tumor suppressor regulates
glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein
degradation. Proc Natl Acad Sci USA. 101:9085–9090. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Sears R, Nuckolls F, Haura E, Taya Y,
Tamai K and Nevins JR: Multiple Ras-dependent phosphorylation
pathways regulate Myc protein stability. Genes Dev. 14:2501–2514.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yeh E, Cunningham M, Arnold H, Chasse D,
Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida
T, et al: A signalling pathway controlling c-Myc degradation that
impacts oncogenic transformation of human cells. Nat Cell Biol.
6:308–318. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Arnold HK, Zhang X, Daniel CJ, Tibbitts D,
Escamilla-Powers J, Farrell A, Tokarz S, Morgan C and Sears RC: The
Axin1 scaffold protein promotes formation of a degradation complex
for c-Myc. EMBO J. 28:500–512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liu L and Eisenman RN: Regulation of c-Myc
protein abundance by a protein phosphatase 2A-glycogen synthase
kinase 3β-negative feedback pathway. Genes Cancer. 3:23–36. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Seo HR, Kim J, Bae S, Soh JW and Lee YS:
Cdk5-mediated phosphorylation of c-Myc on Ser-62 is essential in
transcriptional activation of cyclin B1 by cyclin G1. J Biol Chem.
283:15601–15610. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Sjostrom SK, Finn G, Hahn WC, Rowitch DH
and Kenney AM: The Cdk1 complex plays a prime role in regulating
N-myc phosphorylation and turnover in neural precursors. Dev Cell.
9:327–338. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Junttila MR, Puustinen P, Niemelä M, Ahola
R, Arnold H, Böttzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, et
al: CIP2A inhibits PP2A in human malignancies. Cell. 130:51–62.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bonetti P, Davoli T, Sironi C, Amati B,
Pelicci PG and Colombo E: Nucleophosmin and its AML-associated
mutant regulate c-Myc turnover through Fbw7 gamma. J Cell Biol.
182:19–26. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Welcker M, Orian A, Grim JE, Eisenman RN
and Clurman BE: A nucleolar isoform of the Fbw7 ubiquitin ligase
regulates c-Myc and cell size. Curr Biol. 14:1852–1857. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Chandra S, Priyadarshini R, Madhavan V,
Tikoo S, Hussain M, Mudgal R, Modi P, Srivastava V and Sengupta S:
Enhancement of c-Myc degradation by BLM helicase leads to delayed
tumor initiation. J Cell Sci. 126:3782–3795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Tikoo S and Sengupta S: Time to bloom.
Genome Integr. 1:142010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kim BY, Yang JS, Kwak SY, Zhang XK and Han
YH: NEMO stabilizes c-Myc through direct interaction in the
nucleus. FEBS Lett. 584:4524–4530. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Huang H, Ma L, Li J, Yu Y, Zhang D, Wei J,
Jin H, Xu D, Gao J and Huang C: NF-κB1 inhibits c-Myc protein
degradation through suppression of FBW7 expression. Oncotarget.
5:493–505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Chen J, Shin JH, Zhao R, Phan L, Wang H,
Xue Y, Post SM, Ho Choi H, Chen JS, Wang E, et al: CSN6 drives
carcinogenesis by positively regulating Myc stability. Nat Commun.
5:53842014. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Olive V, Sabio E, Bennett MJ, De Jong CS,
Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A,
et al: A component of the mir-17-92 polycistronic oncomir promotes
oncogene-dependent apoptosis. eLife. 2:e008222013. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Onoyama I, Suzuki A, Matsumoto A, Tomita
K, Katagiri H, Oike Y, Nakayama K and Nakayama KI: Fbxw7 regulates
lipid metabolism and cell fate decisions in the mouse liver. J Clin
Invest. 121:342–354. 2011. View Article : Google Scholar :
|
|
115
|
Tu K, Zheng X, Zan X, Han S, Yao Y and Liu
Q: Evaluation of Fbxw7 expression and its correlation with the
expression of c-Myc, cyclin E and p53 in human hepatocellular
carcinoma. Hepatol Res. 42:904–910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Tu K, Zheng X, Zhou Z, Li C, Zhang J, Gao
J, Yao Y and Liu Q: Recombinant human adenovirus-p53 injection
induced apoptosis in hepatocellular carcinoma cell lines mediated
by p53-Fbxw7 pathway, which controls c-Myc and cyclin E. PLoS One.
8:e685742013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Imura S, Tovuu LO, Utsunomiya T, Morine Y,
Ikemoto T, Arakawa Y, Kanamoto M, Iwahashi S, Saito Y, Takasu C, et
al: The role of Fbxw7 expression in hepatocellular carcinoma and
adjacent non-tumor liver tissue. J Gastroenterol Hepatol.
29:1822–1829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Tien JC and Xu J: Steroid receptor
coactivator-3 as a potential molecular target for cancer therapy.
Expert Opin Ther Targets. 16:1085–1096. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Chen T, Sun Y, Ji P, Kopetz S and Zhang W:
Topoisomerase IIα in chromosome instability and personalized cancer
therapy. Oncogene. Oct 20–2014.Epub ahead of print. View Article : Google Scholar
|
|
120
|
Piccolo S, Dupont S and Cordenonsi M: The
biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev.
94:1287–1312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Lahusen T, Henke RT, Kagan BL, Wellstein A
and Riegel AT: The role and regulation of the nuclear receptor
co-activator AIB1 in breast cancer. Breast Cancer Res Treat.
116:225–237. 2009. View Article : Google Scholar : PubMed/NCBI
|