Introduction
Colorectal cancer (CRC), one of the most prevalent
malignant cancers worldwide, is the third most common cancer in men
and the second in women, accounting for 10.0 and 9.2% of cancer
cases, respectively (1,2). The main cause of the CRC patient death
is metastasis of CRC cells to other organs. The 5-year survival
rate of early stage CRC patients (stage I) is higher than 90%, but
that of advanced stage CRC patients with metastases (stage IV)
decreases to less than 5% (3–5).
Although the CRC incidence rate is gradually decreasing in
developed countries, it is still increasing in numerous developing
countries. Numerous studies are ongoing to elucidate the molecular
mechanisms related to CRC development in order to develop effective
treatments and prognostic markers.
MicroRNAs (miRNAs), endogenous small RNAs of 19–25
nucleotides, regulate translation through mRNA degradation or
translational inhibition by binding to the 3′ untranslated regions
(UTRs) of target mRNAs (6). More
than 50% of miRNAs identified in human cells are encoded in
cancer-associated genomic regions and are deeply involved in human
cancer development (7,8). Among these, miR-330-5p was first
discovered by Weber in 2005 (9),
and several studies have identified miR-330-5p as a
tumor-suppressor in prostate and pancreatic cancers and in CRC cell
lines as well (10–13). In contrast, a study suggested that
miR-330-5p is an oncomiR in glioblastoma cells (14,15).
In any case, these lines of evidence implicate miR-330-5p in human
cancers. Although several target genes are known for miR-330-5p,
the list of its molecular targets is likely to be incomplete and
the mechanisms of its involvement in cancer are not well
understood.
Integrins are a family of transmembrane proteins
that mediate communications between different cells or between
cells and extracellular matrix. They play important roles in tumor
development through regulation of cell proliferation, survival,
migration and invasion (16).
Functional integrins are heterodimeric proteins composed of one α
and one β chain; in humans, 18α and 8β integrin subunits can be
present in any combination (17).
Among the α subunits, α5 (ITGA5) forms a dimer predominantly with
β1 (ITGB1). Integrin α5β1 recognizes the arginine-glycine-aspartic
acid sequence in its ligand fibronectin, which is one of the
proteins organizing the extracellular matrix. Recently, it was
revealed that integrin α5β1 promotes metastasis of CRC cells in the
hepatic microenvironment (18).
Furthermore, ITGA5 was shown to regulate peritoneal dissemination
of ovarian cancer cells and adhesion and invasion of CRC cells,
thus affecting metastasis (19–22).
In the present study, we showed that the level of
miR-330-5p expression was decreased and was inversely related to
that of ITGA5 in the CRC tissue. Our data identified ITGA5 as a new
miR-330-5p target and demonstrated negative regulation of ITGA5
expression by direct binding of miR-330-5p to its 3′UTR. Together
with previous reports, our results suggest that miR-330-5p acts as
an antimetastatic miRNA in CRC by regulating ITGA5 expression.
Materials and methods
Human tissue samples
Human CRC and non-tumor colorectal tissue samples
were obtained from the Seoul St. Mary's Biobank (2013–05).
Colleciton and use of all samples were approved by the
Institutional Review Board (IRB) of the College of Medicine at the
Catholic University of Korea.
Cell culture and transfection
The human CRC cell lines, DLD-1, SNU-C5 and HCT116,
were purchased from the Korean Cell Line Bank (KCLB; Seoul, Korea).
Cell lines were maintained in RPMI-1640 medium (Invitrogen,
Carlsbad, CA, USA) containing 10% fetal bovine serum and 1%
penicillin/streptomycin with 5% CO2 in a 37°C incubator.
miRNAs used in thes present study were purchased from Dharmacon
(Lafayette, CO, USA). CRC cells were seeded onto 60 mm dishes at
70% confluency. After 24 h, miR-330-5p mimic transfection was
performed using DharmaFECT I (Dharmacon) following the
manufacturer's instructions. Transfected cells were harvested 72 h
post-transfection.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cells using the TRIzol
reagent (Invitrogen) according to the manufacturer's instructions.
Complimentary DNA (cDNA) was synthesized using the PrimeScript II
First Strand cDNA Synthesis kit (Takara, Tokyo, Japan) according to
the manufacturer's instructions. The cDNA synthesis for miRNA was
carried out using the Mir-X miRNA First-Strand Synthesis kit
(Clontech, Mountain View, CA, USA). The qRT-PCR for ITGA5
and miR-330-5p was performed using the SYBR Premix Ex Taq II
(Takara) following the manufacturer's instructions. ITGA5
expression levels were normalized against
glyceraldehyde-3-phosphated dehydrogenase (GAPDH) gene expression.
Expression of miR-330-5p was determined by qRT-PCR using the
miR-330-5p primer and mRQ 3′ Primer (both from Clontech) and
normalized against U6 expression. The sequences of the
gene-specific primers are shown in Table I.
 | Table IList of gene-specific primers for
real-time PCR. |
Table I
List of gene-specific primers for
real-time PCR.
| Genes | Accession number | | Sequences | Size (bp) | Tm (°C) |
|---|
| ITGA5 | NM_002205 | F |
5′-CCCCGAGTACCTGATCAAC-3′ | 204 | 60 |
| R |
5′-AGGGATCGAATGTCTGAGCC-3′ |
| GAPDH | NM_002046 | F |
5′-GAGTCAACGGATTTGGTCGT-3′ | 238 | 60 |
| R |
5′-TTGATTTTGGAGGGATCTCG-3′ |
Western blot analysis
Cell lysates were prepared using RIPA buffer [150 mM
sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium
dodecyl sulfate, 50 mM Tris-HCI (pH 8.0)] according to the standard
method. Protein concentrations were determined by the Bradford
assay. Proteins were resolved on 8% sodium dodecyl sulfate
polyacrylamide gel by electrophoresis and transferred to a
nitrocellulose membrane. The membrane was incubated with a rabbit
polyclonal antibody against ITGA5 (1:1,000; Applied Biological
Materials, Vancouver, Canada) and a mouse polyclonal antibody
against β-actin (1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA,
USA). β-actin level was used for normalization.
Plasmid construction
The full length 3′UTR cDNA of ITGA5 was
amplified from total RNAs of DLD-1 cells by RT-PCR following the
standard protocol. The amplified PCR product was cloned into the
pGEM-T Easy vector and the integrity of its sequence was confirmed
by sequencing. The inserted DNA fragment was subcloned into the
psiCHECK-2 vector (Promega, Madison, WI, USA) using the NotI
site (Takara) to generate psiCHECK-2/h_ITGA5 3′UTR. The deletion
constructs, psiCHECK-2/h_ITGA5 3′UTR_deletion I, psiCHECK-2/h_ITGA5
3′UTR_deletion II and psiCHECK-2/h_ITGA5 3′UTR_deletion III, were
generated by site-directed mutagenesis using the QuikChange
Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA).
Primer sequences for plasmid construction are listed in Table II.
 | Table IIList of primers used in the present
study for plasmid construction. |
Table II
List of primers used in the present
study for plasmid construction.
| Name | | Sequences
(5′→3′) | Size (bp) | Tm (°C) |
|---|
| ITGA5 3′UTR | F |
gtcctcccaatttcagactcc | | |
| R |
ctagttctggtcagtggggg | 1032 | 60 |
| ITGA5 3′UTR deletion
I | F |
agctcctctccccagcatacttgaagggcc | | |
| R |
ggcccttcaagtatgctggggagaggagct | 7297 | 55 |
| ITGA5 3′UTR deletion
II | F |
gggcttcttttggatccaaggctgaggacaga | | |
| R |
tctgtcctcagccttggatccaaaagaagccc | 7300 | 55 |
| ITGA5 3′UTR deletion
III | F |
gccctccctgttcgaaaggggagccc | | |
| R |
gggctcccctttcgaacagggagggc | 7299 | 55 |
Luciferase reporter assay
Cells were plated onto 60 mm dishes at 70%
confluency. After 24 h, the cells were co-transfected with
miR-330-5p mimic and 1 μg of the luciferase reporter
construct containing the ITGA5 3′UTR using the Lipofectamine
2000 reagent (Invitrogen). At 72 h post-transfection, the lysates
of the transfected cells were prepared and the luciferase activity
was measured using the Dual-Luciferase Reporter Assay reagent
(Promega) following the protocols recommended by the
manufacturer.
Bioinformatic analysis
Target genes of miR-330-5p were searched using the
TargetScan version 5.2 (http://www.targetscan.org), miRBase (http://www.mirbase.org/) and TargetMiner (http://www.isical.ac.in/~bioinfo_miu/targetminer20.htm)
databases. To determine the relationship between miR-330-5p and
ITGA5, data sets of miRNAs and mRNAs from the NCI60 cell line were
obtained from CellMiner (http://discover.nci.nih.gov/cellminer/). Correlation
was measured between the expressional levels of hsa-miR-330-5p
(MI0000803) and ITGA5 probe set (201389_at) in CRC cell lines.
Statistical analysis
Statistical significance was determined by Student's
t-tests and p-values <0.05 were regarded as statistically
significant.
Results
miR-330-5p is downregulated in CRC
patients
Although miR-330-5p expression has been shown to be
reduced in various tumors in vivo, it has not been studied
in CRC. To investigate the expression of miR-330-5p in CRC, we
compared its expression in human CRC tumors and paired adjacent
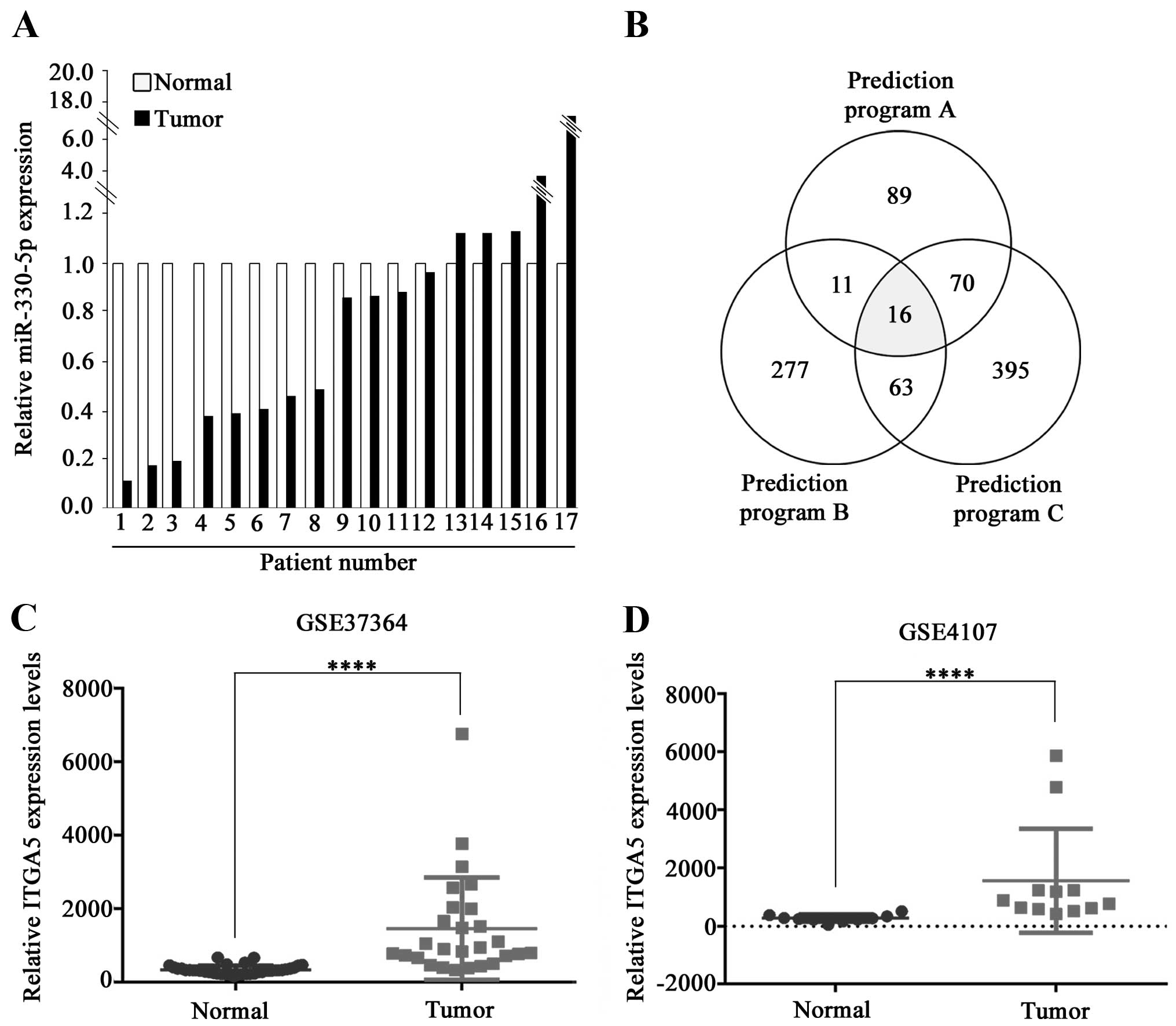
non-tumorous tissues (control) using qRT-PCR. Among the 17 CRC
tumors tested, 12 showed reduced miR-330-5p expression compared to
the control. Eight out of 12 tumors had significantly reduced
miR-330-5p expression (<50%) in comparison with the controls
(Fig. 1A).
Since miRNAs regulate mRNA expression through
recognition of seed-match sequences of the target mRNA, we searched
for putative target genes of miR-330-5p using online software for
prediction of miRNA targets including miRDB, TargetScan and
TargetMiner. Each of these software predicted numerous candidate
target genes and 16 genes were predicted by all three (Fig. 1B). Among them, we focused on
ITGA5 since it plays important roles in the development of
various types of cancers (17,19–23).
Our analysis of the data in the Gene Expression Omnibus (GEO)
database (accession nos. GSE37364 and GSE4107) showed that
ITGA5 expression was significantly increased in the CRC
tumors (Fig. 1C and D).
Expression of miR-330-5p is inversely
correlated with ITGA5 expression in CRC cell lines
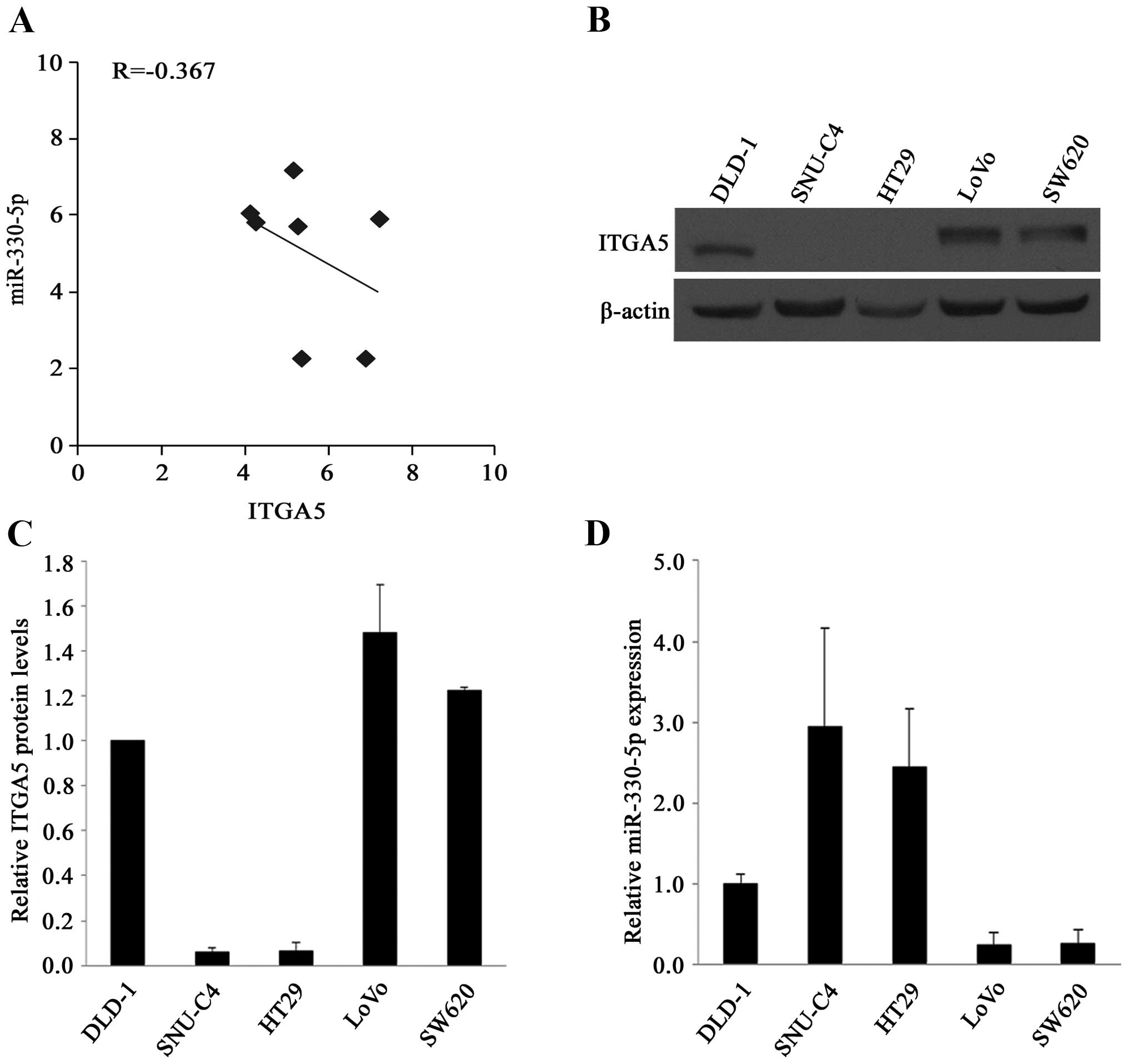
The relationship between the expression of
miR-330-5p and ITGA5 was also investigated using the NCI-60
database of the US National Cancer Institute, which contains mRNA
and miRNA expression data for 60 human cancer cell lines. The
analysis revealed that the expression of miR-330-5p and
ITGA5 was inversely correlated in the 7 CRC cell lines
(R=−0.367) (Fig. 2A).
The relationship between miR-330-5p and ITGA5
protein levels was studied in 5 CRC cell lines. The levels of ITGA5
were determined by western blot analysis and those of miR-330-5p by
qRT-PCR. As shown in Fig. 2B–D, the
inverse relationship was found between the levels of miR-330-5p and
ITGA5 in all 5 cell lines.
miR-330-5p downregulates ITGA5
expression
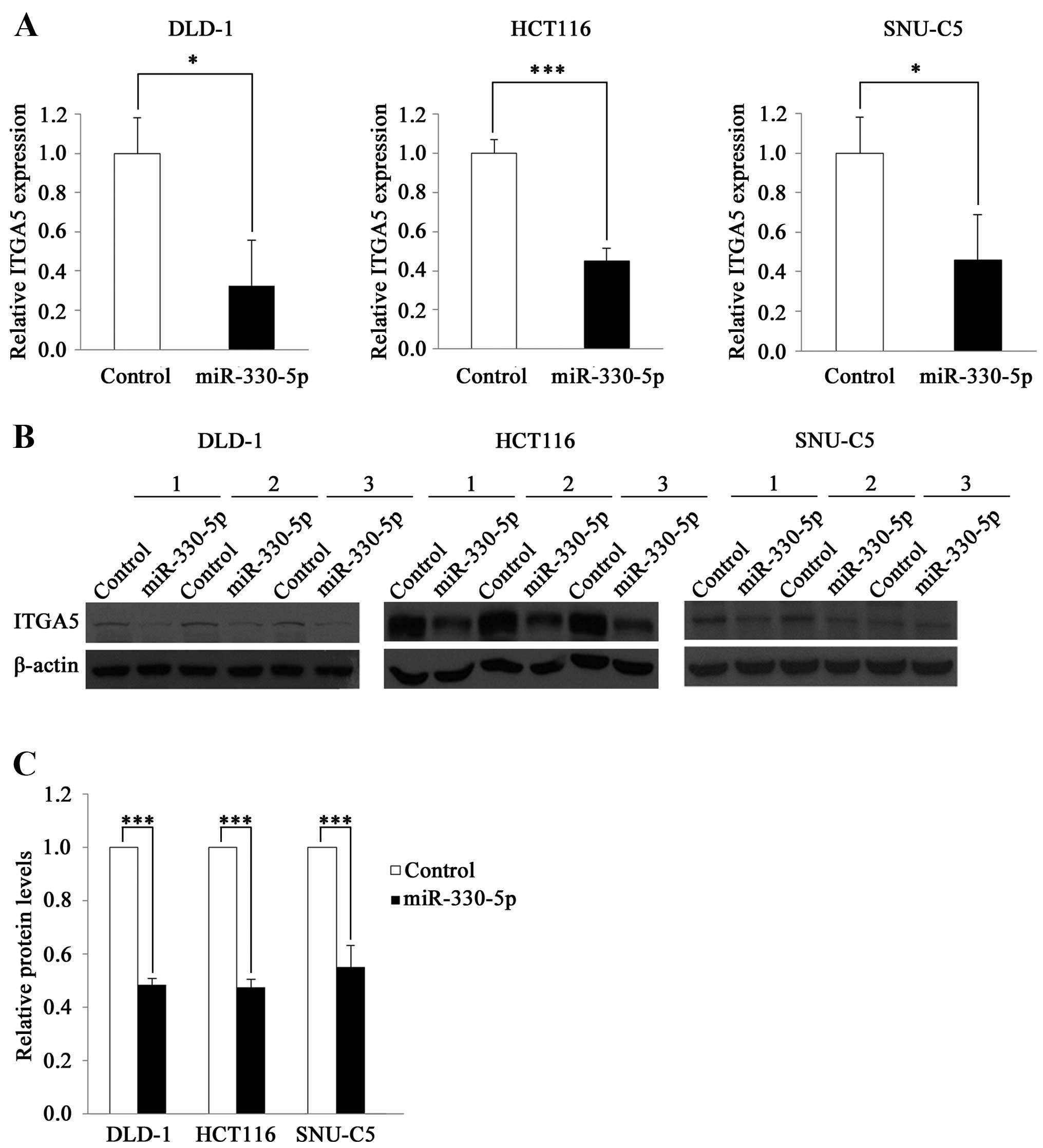
To investigate whether miR-330-5p regulates
ITGA5 expression, we transfected the CRC cell lines DLD-1,
HCT116 and SNU-C5 with a miR-330-5p mimic and measured the levels
of the ITGA5 mRNA. ITGA5 expression was decreased by
>50% in the cell lines transfected with the miR-330-5p mimic
compared to that in cells transfected with the control RNA
(Fig. 3A). Western blot analysis
revealed that the levels of the ITGA5 protein were also reduced by
miR-330-5p: by 51.9% in DLD-1, 52.54% in HCT116 and 59.68% in
SNU-C5 cells (Fig. 3B and C). These
results indicate that miR-330-5p downregulates ITGA5
expression.
miR-330-5p directly targets the 3′UTR of
the ITGA5 mRNA
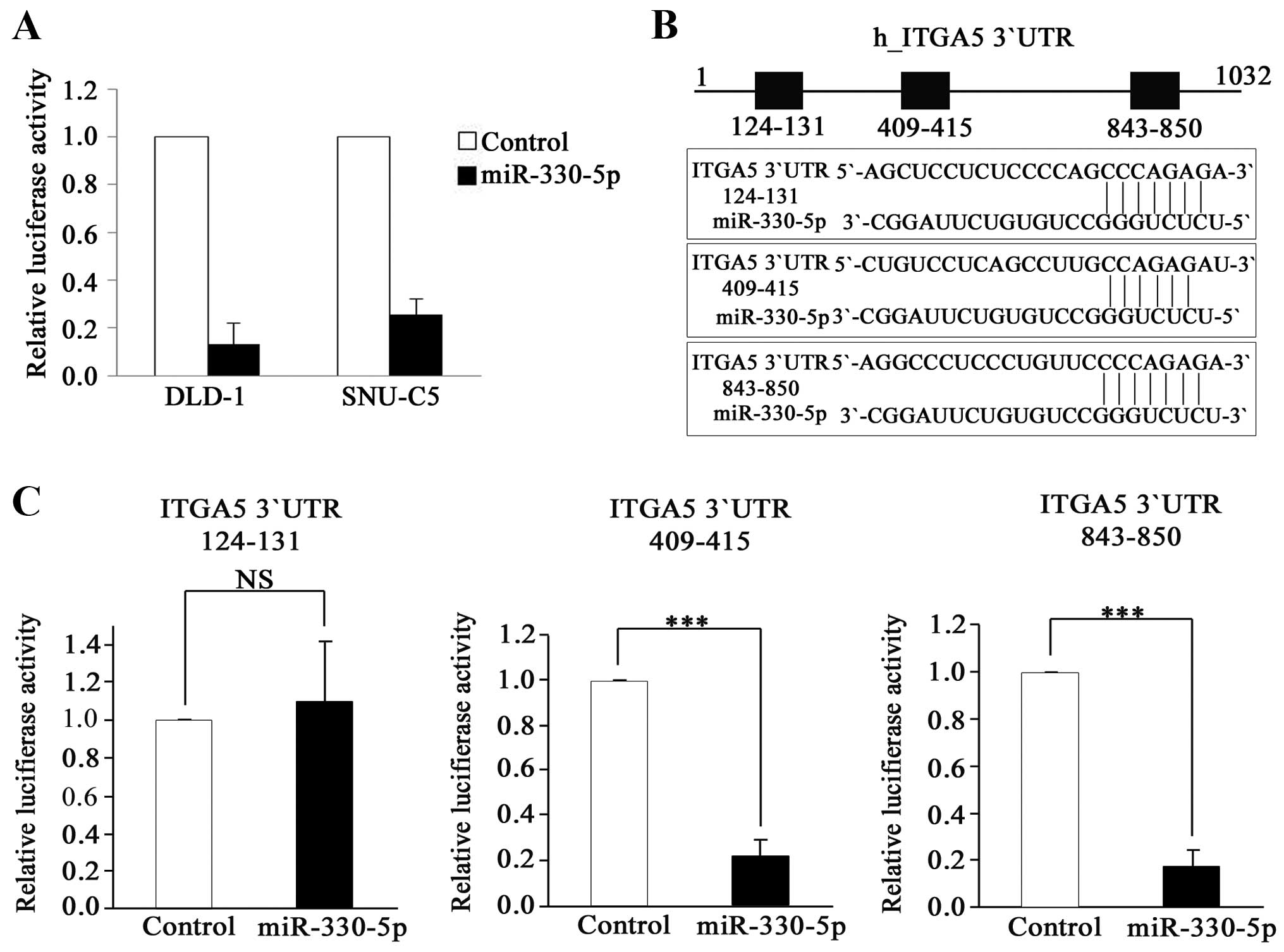
To determine whether miR-330-5p directly binds to
the 3′UTR of the ITGA5 mRNA, we used a heterologous reporter
system. First, we cloned the full-length 3′UTR of ITGA5 into
a reporter plasmid in which luciferase gene expression is affected
by the cloned 3′UTR. The cloned reporter construct was transfected
into the cells along with the miR-330-5p mimic, and luciferase
activity was determined. Luciferase activity was significantly
inhibited by miR-330-5p in both cell lines tested (DLD-1 and
SNU-C5; Fig. 4A). This result
indicated that miR-330-5p directly regulated ITGA5
expression by binding to the 3′UTR of its mRNA. We further
investigated which region of the ITGA5 3′UTR is recognized
by miR-330-5p. Each of the 3 regions predicted as seed matches
(Fig. 4B) was deleted to generate
constructs lacking nucleotides 124-131, 409-415 or 843-850. Each
construct was co-transfected with the miR-330-5p mimic, and
luciferase activity was measured. The construct with the deletion
of nucleotides 124–131 was the only one that did not show a
reduction of luciferase activity by miR-330-5p, indicating that
this region is the target of miR-330-5p (Fig. 4C). This result strongly suggests
that miR-330-5p regulates ITGA5 expression by directly binding to
nucleotides 124–131 of the ITGA5 3′UTR.
Discussion
Accumulating lines of evidence have led to the view
that microRNAs (miRNAs) play important roles in the progression of
human cancers through modulation of proliferation, survival,
metastasis and invasion of the cancer cells. Therefore, the
functions of miRNAs in cancer have been intensively investigated;
this research facilitates the development of miRNA-based diagnosis,
prognosis and therapy (24).
Several studies have identified the function of
miR-330-5p in cancer progression. miR-330-5p was shown to play a
role as an oncomiR by targeting the SH3-domain GRB2-like 2
(SH3GL2) gene in glioblastoma stem cells (14). In contrast, other studies
demonstrated that miR-330-5p inhibits cell motility by targeting
the Sp1 transcription factor (SP1) and induces apoptosis
through E2F transcription factor 1 (E2F1)-mediated
suppression of Akt phosphorylation in prostate cancer (10,11).
Thus, the role of miR-330-5p in tumorigenesis is controversial. A
recent study showed that miR-330-5p inhibited proliferation of CRC
cell lines by downregulating cell division cycle 42 (CDC42)
expression and revealed that miR-330-5p overexpression induced
apoptosis and reduced tumor weight in vivo (13). In the present study, we showed that
miR-330-5p expression was downregulated in tumor compared to
adjacent normal tissues in ~70% of the investigated CRC patients,
raising a possibility of miR-330-5p being a tumor-suppressor in
CRC.
We found an inverse relationship between the
expression of miR-330-5p and ITGA5 in CRC. A similar relationship
(R=−0.54) was also observed in leukemia cell lines (NCI-60
database; data not shown). In contrast, a positive correlation was
found in several other cancer cell lines including neuronal,
non-small cell lung and renal cancer (NCI-60; data not shown).
These results suggest that the relationship between miR-330-5p and
ITGA5 is cancer type-specific.
ITGA5, the newly found target of miR-330-5p, was
found to promote the development of various types of cancers.
Downregulation of ITGA5 inhibited peritoneal dissemination of
ovarian cancer cells; upregulation of ITGA5 promoted adhesion,
invasion and epithelial-mesenchymal transition of CRC cells
(19-22). Furthermore, ITGA5 in a complex with
ITGB1 (integrin α5β1) promoted invasive migration and metastasis of
cervical cancer cells (25) and
regulated ionizing radiation-induced adhesion of breast cancer
cells (26). Thus, there is enough
evidence to support the notion that integrin α5β1 plays important
roles in the development of various human cancers.
Since ITGB1 forms a heterodimer with ITGA5, we
investigated the relationship between the expression of ITGB1 and
miR-330-5p in CRC using public datasets, but found no correlation
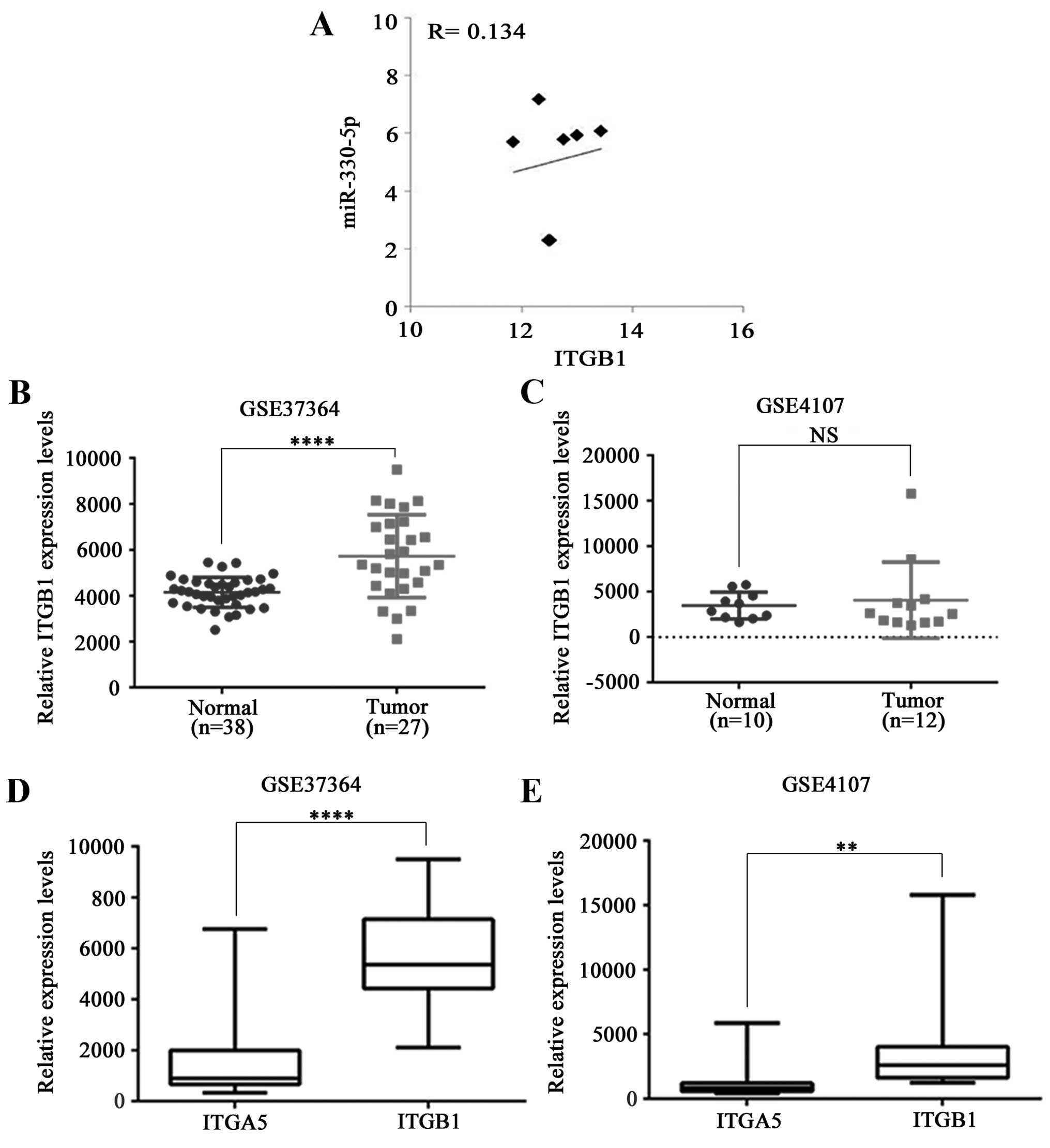
in the 7 CRC cell lines (NCI-60 data; Fig. 5A), and the software for the miRNA
target did not predict ITGB1 as a miR-330-5p target. Based
on these data, ITGB1 seems not to be regulated by miR-330-5p in
CRC.
We further investigated ITGB1 expression using the
same GEO data (GSE37364 and GSE4107) as for the analysis of ITGA5
expression in CRC, and found no consistency between these two
datasets. Whereas ITGB1 mRNA level was slightly higher in
the CRC tumors compared to that of the normal tissue in GSE37364,
there was no difference in the GSE4170 dataset (Fig. 5B and C). At this point, it is not
clear whether ITGB1 expression is related to the progression of
CRC. Whether ITGB1 is in excess, the level of ITGA5 may be limiting
for α5β1 formation, which may explain the apparent irrelevance of
the ITGB1 level. Thus, we compared expression of ITGA5 and ITGB1 in
the same datasets and found 6.3- (ranging from 1.0- to 10.2-fold in
GSE37364) and 3.4- (ranging from 1.3- to 8.0-fold in GSE4107) fold
higher expression of ITGB1 on average (Fig. 5D and E). A similar expression
pattern was noted in lung cancer (27). Obviously further study is required
to understand its relevance to CRC development.
In conclusion, we found that miR-330-5p was
significantly downregulated in human CRC tumors, which led to the
upregulation of ITGA5, a direct target of miR-330-5p. Our findings
suggest that downregulation of miR-330-5p may stimulate the
metastasis and invasion of CRC cells by directly targeting ITGA5.
These results improve our understanding of CRC development and may
be useful for future clinical applications.
Acknowledgments
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education, Science and Technology
(2012R1A5A2047939)
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chu E: Colorectal cancer (CRC) continues
to be a major public health problem in the United States and
throughout the world. Cancer J. 16:1952010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Din FV, Theodoratou E, Farrington SM,
Tenesa A, Barnetson RA, Cetnarskyj R, Stark L, Porteous ME,
Campbell H and Dunlop MG: Effect of aspirin and NSAIDs on risk and
survival from colorectal cancer. Gut. 59:1670–1679. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Cutsem E, Cervantes A and Nordlinger
B: Metastatic colorectal cancer: ESMO Clinical Practice Guidelines
for diagnosis, treatment and follow-up. Ann Oncol. 25(Suppl 3):
iii1–iii9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong Y, Yu J and Ng SS: MicroRNA
dysregulation as a prognostic biomarker in colorectal cancer.
Cancer Manag Res. 6:405–422. 2014.PubMed/NCBI
|
|
7
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar :
|
|
9
|
Weber MJ: New human and mouse microRNA
genes found by homology search. FEBS J. 272:59–73. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT,
Goan YG and Lu PJ: MicroRNA-330 acts as tumor suppressor and
induces apoptosis of prostate cancer cells through E2F1-mediated
suppression of Akt phosphorylation. Oncogene. 28:3360–3370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mao Y, Chen H, Lin Y, Xu X, Hu Z, Zhu Y,
Wu J, Xu X, Zheng X and Xie L: microRNA-330 inhibits cell motility
by downregulating Sp1 in prostate cancer cells. Oncol Rep.
30:327–333. 2013.PubMed/NCBI
|
|
12
|
Tréhoux S, Lahdaoui F, Delpu Y, Renaud F,
Leteurtre E, Torrisani J, Jonckheere N and Van Seuningen I:
Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by
targeting the MUC1 mucin in pancreatic cancer cells. Biochim
Biophys Acta. 1853:2392–2403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Zhu X, Xu W, Wang D and Yan J:
miR-330 regulates the proliferation of colorectal cancer cells by
targeting Cdc42. Biochem Biophys Res Commun. 431:560–565. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z
and Liu Y: MicroRNA-330 is an oncogenic factor in glioblastoma
cells by regulating SH3GL2 gene. PLoS One. 7:e460102012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yao Y, Xue Y, Ma J, Shang C, Wang P, Liu
L, Liu W, Li Z, Qu S, Li Z, et al: MiR-330-mediated regulation of
SH3GL2 expression enhances malignant behaviors of glioblastoma stem
cells by activating ERK and PI3K/AKT signaling pathways. PLoS One.
9:e950602014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guan JL: Role of focal adhesion kinase in
integrin signaling. Int J Biochem Cell Biol. 29:1085–1096. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Walter RB, Laszlo GS, Alonzo TA, Gerbing
RB, Levy S, Fitzgibbon MP, Gudgeon CJ, Ries RE, Harrington KH,
Raimondi SC, et al: Significance of expression of ITGA5 and its
splice variants in acute myeloid leukemia: A report from the
Children's Oncology Group. Am J Hematol. 88:694–702. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pelillo C, Bergamo A, Mollica H, Bestagno
M and Sava G: Colorectal cancer metastases settle in the hepatic
microenvironment through α5β1 integrin. J Cell Biochem.
116:2385–2396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murillo CA, Rychahou PG and Evers BM:
Inhibition of alpha5 integrin decreases PI3K activation and cell
adhesion of human colon cancers. Surgery. 136:143–149. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nam EH, Lee Y, Moon B, Lee JW and Kim S:
Twist1 and AP-1 cooperatively upregulate integrin α5 expression to
induce invasion and the epithelial-mesenchymal transition.
Carcinogenesis. 36:327–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nam EH, Lee Y, Zhao XF, Park YK, Lee JW
and Kim S: ZEB2-Sp1 cooperation induces invasion by upregulating
cadherin-11 and integrin α5 expression. Carcinogenesis. 35:302–314.
2014. View Article : Google Scholar
|
|
22
|
Ohyagi-Hara C, Sawada K, Kamiura S, Tomita
Y, Isobe A, Hashimoto K, Kinose Y, Mabuchi S, Hisamatsu T,
Takahashi T, et al: miR-92a inhibits peritoneal dissemination of
ovarian cancer cells by inhibiting integrin α5 expression. Am J
Pathol. 182:1876–1889. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Goodman SL and Picard M: Integrins as
therapeutic targets. Trends Pharmacol Sci. 33:405–412. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu D, Zhang XX, Wan DY, Xi BX, Ma D, Wang
H and Gao QL: Sine oculis homeobox homolog 1 promotes α5β1-mediated
invasive migration and metastasis of cervical cancer cells. Biochem
Biophys Res Commun. 446:549–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee SH, Cheng H, Yuan Y and Wu S:
Regulation of ionizing radiation-induced adhesion of breast cancer
cells to fibronectin by alpha5beta1 integrin. Radiat Res.
181:650–658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dingemans AM, van den Boogaart V, Vosse
BA, van Suylen RJ, Griffioen AW and Thijssen VL: Integrin
expression profiling identifies integrin alpha5 and beta1 as
prognostic factors in early stage non-small cell lung cancer. Mol
Cancer. 9:1522010. View Article : Google Scholar : PubMed/NCBI
|