Introduction
Being a frequent pathological process,
atherosclerosis underlies adverse cardiovascular events, such as
coronary artery disorder, ischemic gangrene, abdominal aortic
aneurisms and stroke (1). Clinical
complications derived from atherosclerosis, including stroke and
myocardial infarction are contributors to majority of the morbidity
and mortality of cardiovascular disorders worldwide. There is still
an unfulfilled need for non-statin drugs for patients with normal
levels of LDL-cholesterol and/or who are intolerant to statins or
are maximized on current therapy although statin therapy has been
demonstrated to significantly decrease mortality of cardiovascular
diseases in patients with hypercholesterolemia (2).
OSBPL1A affects the target known as late endocytic
compartments (LE) through the endoplasmic reticulum (ER) and the
small GTPase Rab7 via VAMP-related proteins (VAPA and -B) (3). OSBPL1A acts as a sterol-specific
regulator in the interactions between LE and ER membranes. OSBPL1A
regulates the subcellular distribution and tethering and motility
of the endosomes via the regulation of dynein/dynactin and
homotypic fusion and protein sorting (HOPS) complexes and by
linking the bridge between the ER and LE (4). The effect of OSBPL1A in lipid
metabolism is not clear. In low-density lipoprotein receptor
knock-out animals, overexpression of human OSBPL1A in mouse
macrophages enhanced atherogenesis and disturbed cholesterol efflux
to HDL (5).
As a family of small non-coding RNAs, microRNAs
(miRNAs) bind to the 3′ untranslated region (UTR) of target mRNAs
to play a post-transcriptional regulatory role in gene expression.
This causes the suppression of translation or the cleavage of the
target mRNA by the Ago2 ribonuclease in the RNA-triggered silencing
complex (RISC) (6). In addition,
miRNAs can play their roles by regulation of the coordination
between target mRNAs and effector instead of functioning as
modulators of specific mRNAs (7).
Deregulation of potential altered gene expression and miRNA
expression may lead to the occurrence of cancerous phenotypes
(8). Many microRNAs have been
reported as having a role in the process of atherosclerosis
(9). Here we highlight circulating
miRNAs transported by LDL or HDL particles and briefly discusses
circulating miRNAs identified to be implicated in intercellular
interaction within atherosclerosis (10,11).
Circulating miRNA profiles have been demonstrated to vary in
patients with hyperlipidemia and atherosclerosis (12,13).
Moreover, it was demonstrated that, compared with other portions of
the genome, SNPs are less common in miRNAs or their target sites
(14). This negative selection of
sequence variations in miRNAs strengthens their significance for
key cellular processes such as the modulation of gene expression.
It is conceivable that the impact of miR-SNPs can be explained in
different scenarios. On the one hand, SNPs in a miRNA-coding
sequence might impact the expression of an array of different
genes, for instance, because of a compromised maturation process or
processing of the miRNA. On the other hand, SNPs in target sites
can either regulate existing binding sites or build new binding
sites to play their roles in one specific target molecule or in a
few.
OSBPL1A has been demonstrated to play an important
role in the metabolism of lipid, especially HDL, and it has been
also shown that miR-499a targeted OSBPL1A (15,16).
One polymorphism in the pri-miR-499a has been reported to be able
to compromise the processing the pri-miRNA and lower its production
level (15). In this study, we
validated the miR-499a/OSBPL1A involvement in HepG2 cells, and
determined the correlation between the polymorphism of miR-499a
rs3746444 and the expression of miR-499a, its target gene as well
as its association with HLD level.
Materials and methods
Participants
Forty-eight subjects without health problems were
collected at China-Japan Union Hospital of Jilin University, and
peripheral blood samples were collected from these 46 subjects, and
stored at −80°C for prolonged storage for future use. The protocol
of the study was approved by the Ethics Committee of China-Japan
Union Hospital of Jilin University and written informed consents
were obtained from all participants prior to the study.
Serum RNA isolation and real-time
PCR
Serum samples frozen were thawed at room
temperature, and Qiagen® miRNeasy kit
(Qiagen® GmbH, Hilden, Germany) was utilized to extract
total RNA from 500 µl serum collected from the subjects according
to the manufacturer's instructions. The PrimerScript RT reagent kit
(Takara, Dalian, China) was used to perform the reverse
transcription in 20 µl of reaction volume, and the reaction was
carried out at 37°C for 25 min, then maintained at 85°C for 5 sec.
ABI7500 (Applied Biosystems, Foster City, CA, USA) with SYBR Premix
Ex Taq™ II (Takara) was used to perform real-time polymerase chain
reaction with the cDNA synthesized. The reaction was carried out at
95°C for 30 sec (initial denaturation), 40 cycles of 95°C for 5 sec
and 60°C for 30 sec (amplification). The internal control included
small nuclear RNA U6 and β-actin. The cycle threshold (Ct) value
was used to express and analyze the relative miRNA-481 levels.
2−∆∆CT (the comparative CT method) was used to present
the expression of OSBPL1A gene and miR-499a. All reactions were
performed in triplicate.
RNA isolation and real-time PCR
TRIzol® reagent (Invitrogen, NY, USA) was
used to isolate total from HepG2 cells line and tissue samples
according to the manufacturer's instructions. The PrimerScript RT
reagent kit (Takara) was used to perform the reverse transcription
in 20 µl of reaction volume, and the reaction was carried out at
37°C for 25 min, then maintained at 85°C for 5 sec. ABI7 500
(Applied Biosystems) with Premix Ex Taq II (Takara) was used to
perform real-time polymerase chain reaction with the cDNA
synthesized. The reaction was carried out at 95°C for 30 sec
(initial denaturation), 40 cycles of 95°C for 5 sec and 60°C for 30
sec (amplification). The internal controls were the small nuclear
RNA U6 and β-actin. The cycle threshold (Ct) value was used to
express and analyze the relative miR-499a levels. 2−∆∆CT
was used to present the expression of OSBPL1A gene and miR-499a.
All reactions were performed in triplicate.
Cell culture and transfection
RPMI-1640 (Roswell Park Memorial Institute) (Gibco,
Grand Island, NY, USA) contained 10% FBS (fetal bovine serum)
(Gibco), 2 mM glutamine (Sigma, USA) and 1% streptomycin/penicillin
was used to incubate the HepG2 cells at 37°C with 5%
CO2. miR-499a mimics and inhibitor were synthesized from
RiboBio Co. (Guangzhou, China). In brief, the HepG2 cells seeded in
48-well plates at a concentration of 1×105 per well for
12 h, Lipofectamine 2000 (Invitrogen, CA, USA) was used to
transfect the cells with miR-499a mimics or inhibitor and OSBPL1A
siRNA according to the manufacturer's instructions. Each was run
three times.
Luciferase assay
PCR was performed to amplify OSBPL1A 3′UTR with
putative miR-499a binding site from human genomic DNA purchased
from Novagen (Madison, WI, USA), and mutation was introduced by
using site-directed mutagenesis. Both wild-type and mutant 3′UTR of
OSBPL1A were inserted into dual luciferase reporter vector
(Promega, Madison, WI, USA) individually. Lipofectamine 2000
(Invitrogen) was used to co-transfect the HepG2 cells with 100 nM
wild-type/mutant type reporter construct and 50 nM miR-499a mimic
according to the manufacturer's instructions. Forty-eight hours
after transfection, dual luciferase assay (Promega) was utilized to
measure ratio of Renilla luciferase activity to firefly luciferase
activity according to the manufacturer's instructions. All
reactions were carried out three times.
Western blot analysis
The HepG2 cells were transfected with miR-499a
mimics or inhibitor, and the cells were harvested 48 h after
transfection, and ice-cold PBS buffer was used to wash the cells
three times. RIPA (radioimmunoprecipitation assay) lysis buffer
(Upstate Biotechnology, Lake Placid, NY, USA) was used to extract
the protein from the HepG2 cells and tissue sample according to the
manufacturer's instructions. The protein assay reagents (Bio-Rad
Laboratories, Hercules, CA, USA) was used to determine the protein
concentration following the standard protocol. SDS-PAGE was used to
separate the proteins, and then electro-transferred to a
nitrocellulose membrane (Bio-Rad Laboratories), and
Odyssey® blocking buffer (LI-COR Biosciences, Lincoln,
NE, USA) was used to treat the membranes at room temperature for 60
min avoiding unspecific binding, then monoclonal mouse anti-β-actin
antibody (1:10,000, Cell Signaling Technology, Inc., Beverly, MA,
USA) and polyclonal rabbit anti-OSBPL1A antibody (1:5,000) (Cell
Signaling Technology) were carried out to detect the target protein
for 12 h at 4°C, and the horseradish peroxidase [(HRP)-conjugated
rabbit anti-goat IgG secondary antibodies (1:15,000, Cell Signaling
Technology)] were used to visualize the bound antibodies for 2 h at
room temperature. Odyssey CLx imaging system, model: Ody-3086
(LI-COR Inc.)
Statistical analysis
Each test was performed in triplicate. One-way ANOVA
and Student's t-test were utilized to determine statistical
significance, and Dunnett's multiple comparison tests was also
performed to measure the statistical significance, and SigmaStat
3.1 software (Sigma-Aldrich, St. Louis, MO, USA) was utilized to
perform the analyses. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-499a directly targets osbpl1a
Bioinformatics algorithms including RNAhybrid and
TargetScan were utilized to predict the miR-499a target gene. Based
on the results of algorithms above, we predicted osbpl1a might be a
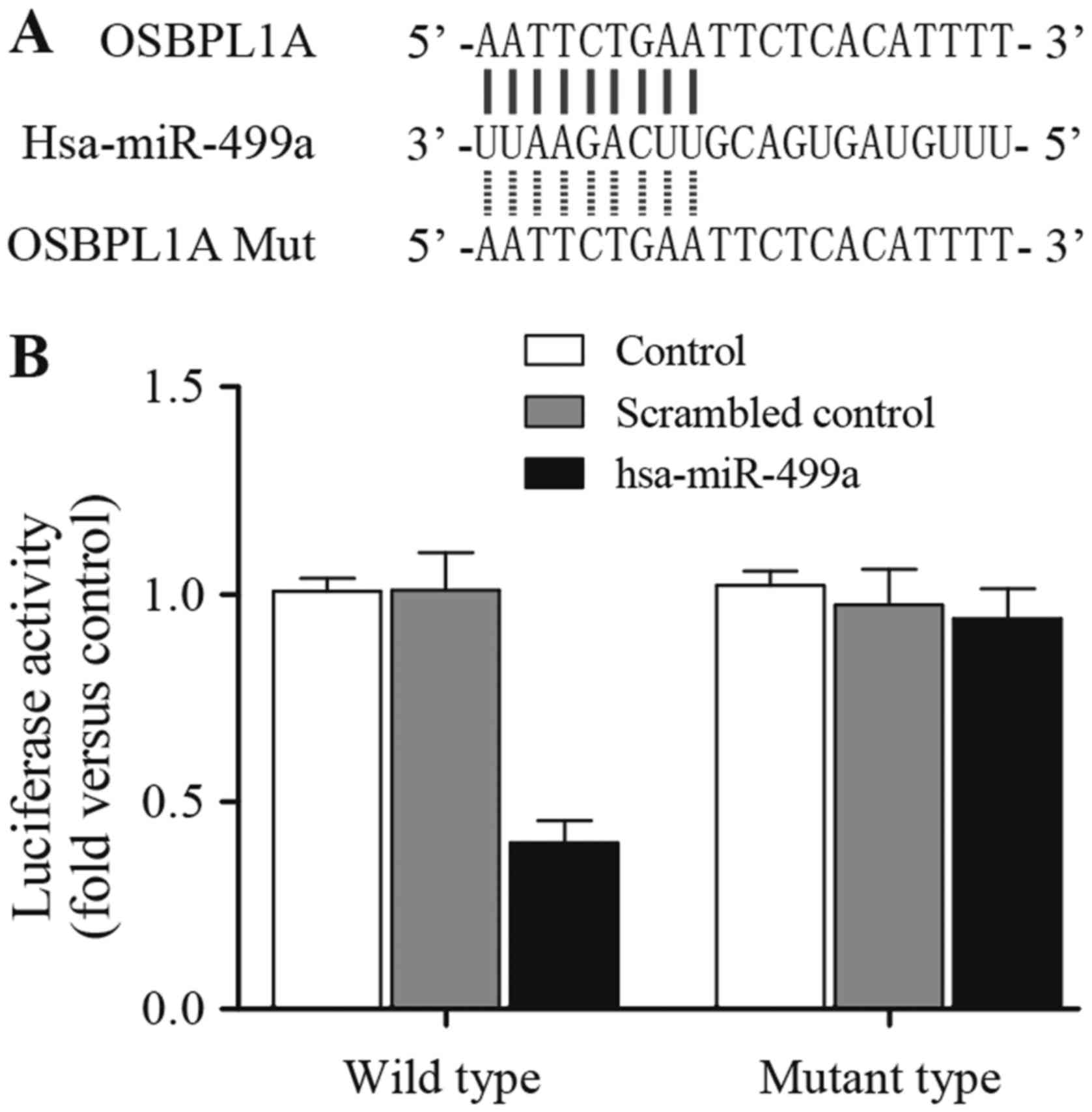
possible target gene of miR-499a with a complementary seed region
of miR-499a (Fig. 1A), then we
mutated the seed region using site-directed mutagenesis, and
obtained mutant type osbpl1a 3′UTR. To further confirm osbpl1a was
a candidate gene of miR-499a, we then conducted luciferase assay,
and subcloned wild or mutant osbpl1a 3′UTR into luciferase reporter
which located directly downstream of luciferase gene. Then cells
co-transfected with luciferase reporter carried wild or mutant
osbpl1a 3′UTR and miR-499a or scramble control. As shown in
Fig. 1B, the luciferase activity of
cells co-transfected with wild osbpl1a 3′UTR and miR-499a was
substantial downregulated compared to that in scramble control,
while transfection of mutant osbpl1a 3′UTR abolished the inhibitory
effect of miR-499a, indicated that miR-499a directly target osbpl1a
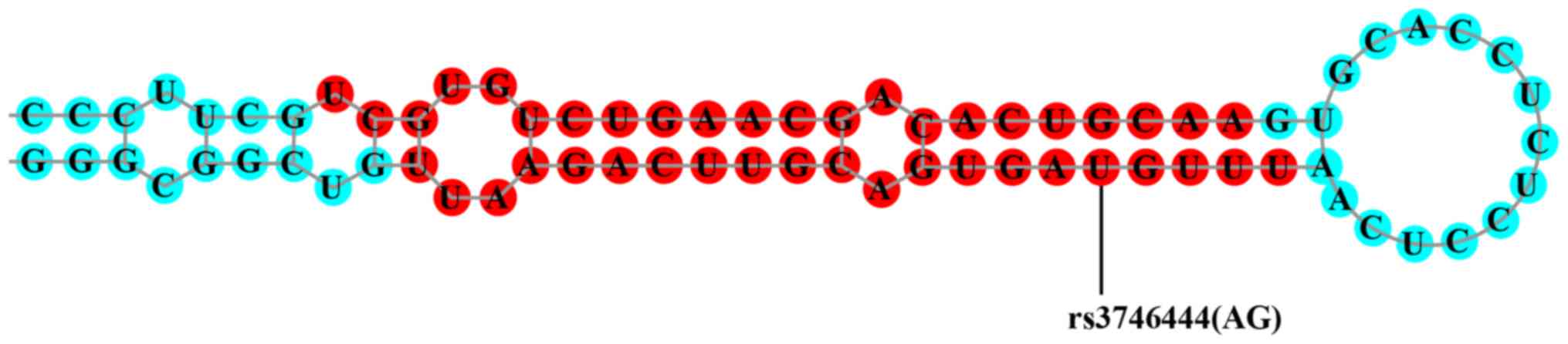
3′UTR, and repressed osbpl1a expression. As shown in Fig. 2, a single nucleotide polymorphism
(rs3746444) in the miR-499a compromises the production of the
miRNA.
miR-499a influences osbpl1a
expression
Based on the results of in silico analysis
and luciferase assay, osbpl1a was a virtual target gene of
miR-499a, to further confirm the miRNA-mRNA regulatory relationship
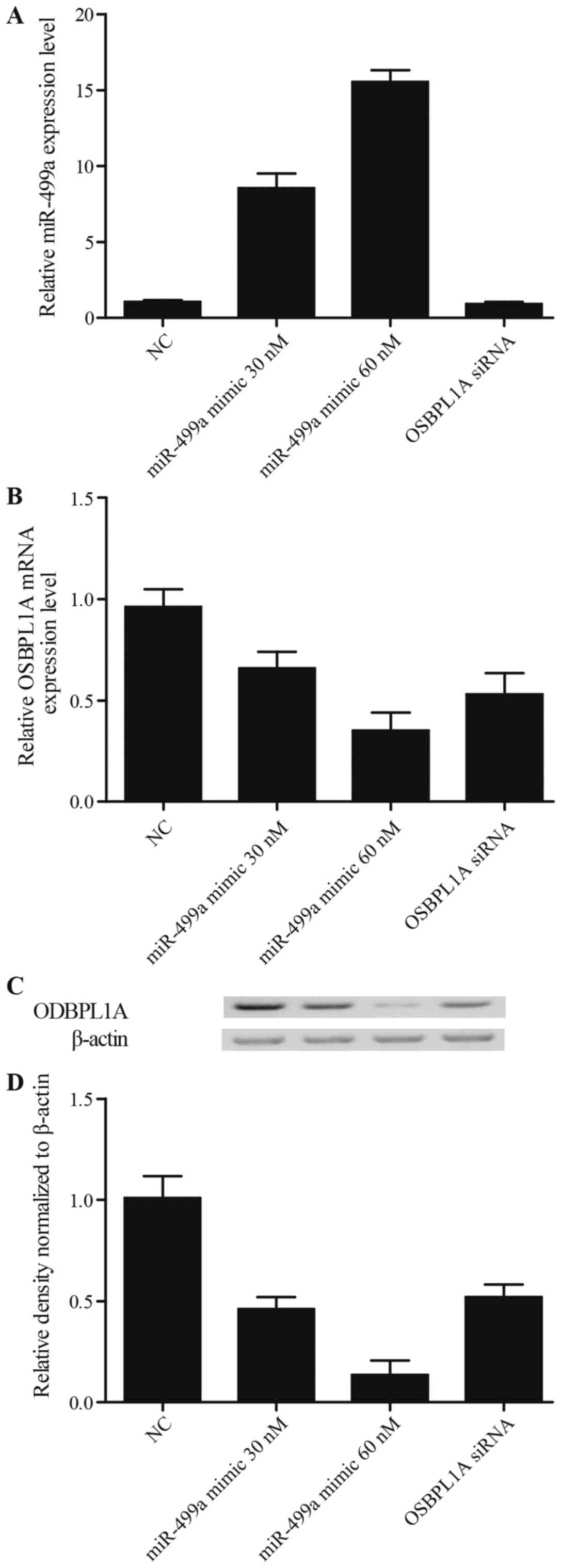
between miR-499a and osbpl1a, we transfected HepG2 cells with
various concentrations of miR-499a mimic (30 and 60 nM),
anti-miR-499a mimic (30 and 60 nM), osbpl1a siRNA and negative
control (NG), β-actin was used as internal control. Then miR-499a
mRNA, osbpl1a mRNA and protein were examined using real-time PCR
and western blot analysis. As shown in Fig. 3A, osbpl1a siRNA did not affect
miR-499a level compared with NC group, the miR-499a level of cells
treated with 30 nM miR-499a mimics were apparently higher than the
scramble control, and those of the cells treated with 60 nM
miR-499a mimics were even higher than the 30 nM treatment group. As
shown in Fig. 3B-D, both osbpl1a
mRNA (Fig. 3B) and protein
(Fig. 3C and D) levels of cells
transfected with miR-449a (30 and 60 nM) or osbpl1a siRNA were
markedly reduced in comparison with NC groups, furthermore the
inhibitory effect of either 30 nM miR-499a mimic or 60 nM miR-499a
mimic on osbpl1a expression was comparable with that of osbpl1a
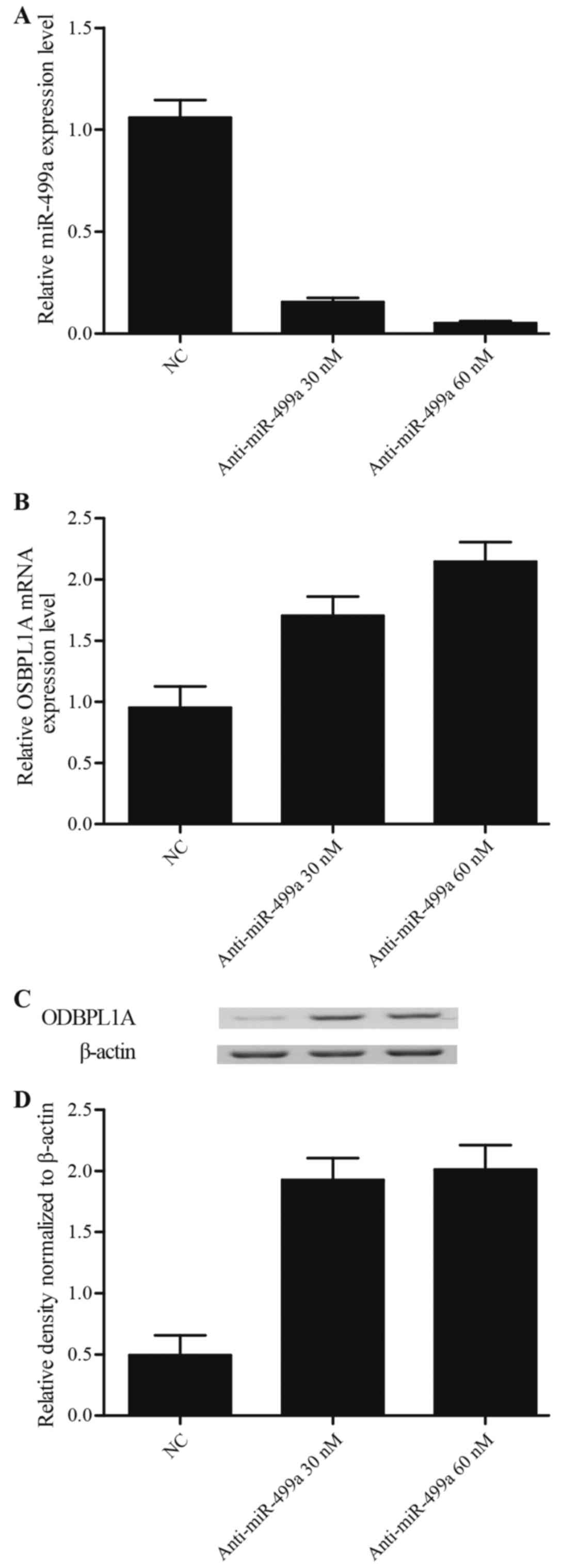
siRNA. By contrast, as shown in Fig.
4A, the miR-499a expression of cells treated with 30 nM
anti-miR-499a mimics and 60 nM anti-miR-499a mimics were attenuated
compared to the scramble control, and the suppression effect of 60
nM anti-miR-499a mimic on miR-499a expression level was much
stronger than 30 nM anti-miR-499a mimic groups. As shown in
Fig. 4B-D, both mRNA (Fig. 4B) and protein (Fig. 4C and D) levels of osbpl1a in cells
transfected with anti-miR-449a (30 and 60 nM) were notably
upregulated in comparison with NC groups, furthermore, the osbpl1a
mRNA (Fig. 4B) and protein
(Fig. 4C and D) in 30 nM miR-499a
mimic treatment group exhibited no obvious difference with 60 nM
miR-499a mimic treatment group, suggested that a
concentration-dependent effect of miR-499a on the miR-499a
expression, and miR-449a negatively regulated osbpl1a expression in
a concentration-independent manner.
The expression of miR-499a and HDL
level in different genotype groups
Forty-six volunteers diagnosed with atherosclerosis
took part in our study. Also, information of participants, such as
sex, age and HDL level were collected. To confirm the effect of
rs3746444 polymorphism on the expression of miR-499a, we divided
the 46 participants into three groups by genotypes: AA, AG and GG,
and the frequency distribution of the AA, AG, GG genotypes was 22,
18 and 6, respectively. Real-time quantitative PCR was performed to
determine the level of miR-499a in serum derived from the 46
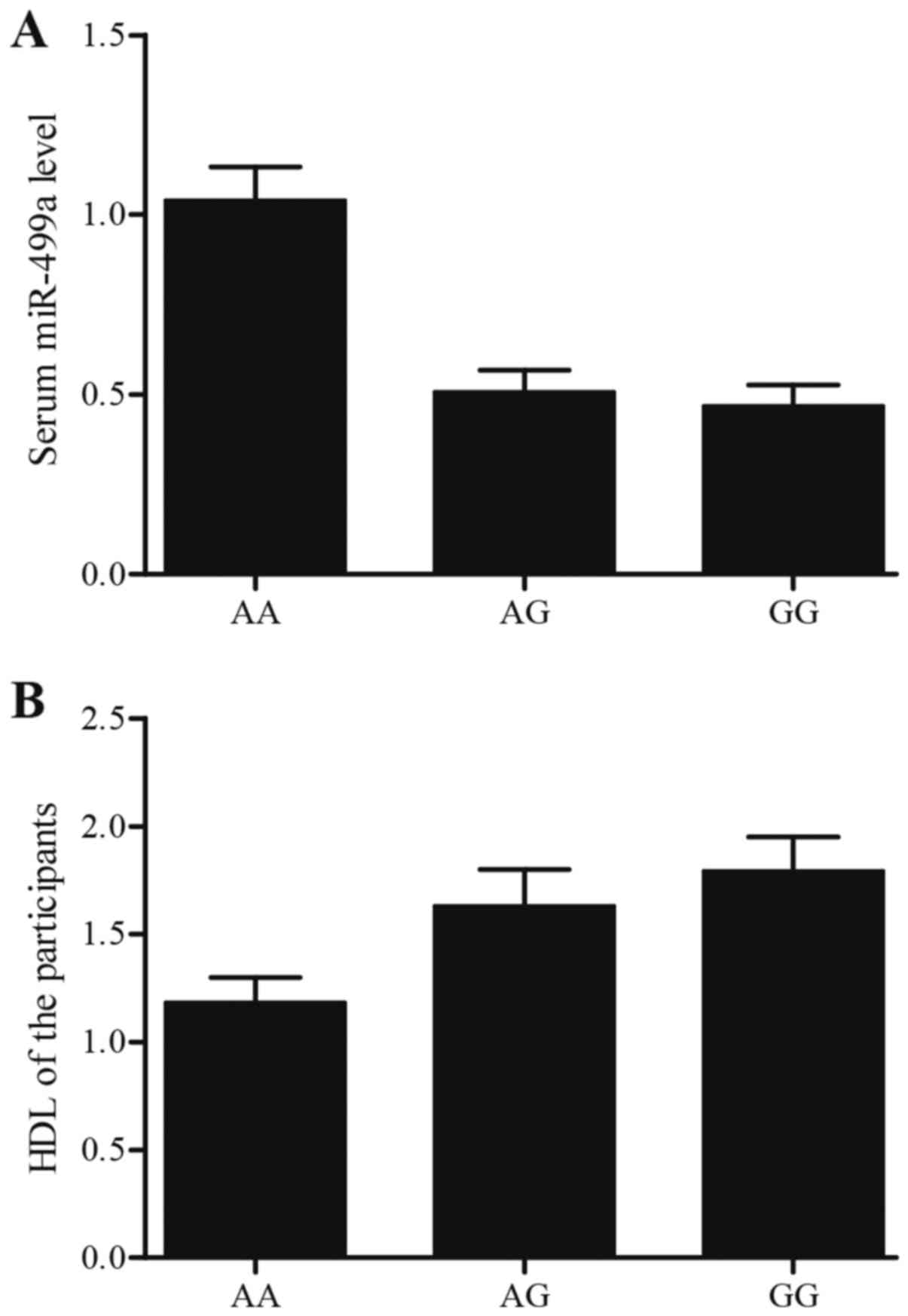
participants, as shown in Fig. 5A,
miR-499a was overexpressed in AA group compared with AG and GG
groups, and miR-499a level in GA group and GG group was similar.
The result validated that the influence of rs3746444 A allele on
expression level of miR-499a represented a recessive pattern in
high-grade group. One-way ANOVA and Student's t-test were utilized
to determine the difference on sex, age and HDL level among AA, AG
and GG groups, as shown in Fig. 5B,
HDL level in AA group was much lower than AG and GG groups, and sex
or age in AA, AG and GG comparable with each other.
Discussion
Increasing data indicate that large amounts of MVs
are released in human atherosclerotic plaques following the
activation or apoptosis cells. Although the stimuli that mediate
miRNA secretion are unclear (17).
In addition, it has been shown that plaque MVs anchor on
non-activated ECs and account for the development and progression
of atherosclerosis (18).
Particularly, patients with atherosclerosis have high levels of
miR-499a-3p and miR-135b-5p (19).
Hence, ECs might be a better option compared with VSMCs, because
ECs have minimal expression of miR-499a-3p and miR-135-5p, and
endothelial dysfunction is especially a driver in the mediation and
progression of atherosclerosis (20). miR-499a is known as a potential AMI
biomarker because the cardiac muscle has highly specific expression
of miR-499a (21). The significant
reduction (2.1-fold) in the production of miR-499a that we found in
the AMI samples was evidenced by earlier studies that miR-499 is
significantly increased in the serum of patients with AMI
(indicator of release from the cardiac tissue) 4–12 h following AMI
and is significantly reduced in the zone with cardiac infarction in
an AMI mouse model (22,23). In this study, we used online miRNA
target prediction tools to search the target gene of miR-499a, and
found that osbpl1a might be a possible target gene of miR-499a with
a complementary seed region of miR-499a located within osbpl1a
3′UTR. Next, we conducted luciferase reporter assay to validate
osbpl1a as a direct target gene of miR-499a.
As a member of a family of sterol sensors, the gene
known as OSBPL1A could have an effect on lipid metabolism (24). It is thought that epigenetic
modification has another critical role involving nutrient sensing
(25). OSBPL1A is present at
20 kb from the imprinted gene IMPACT, however, mouse brain
does not show an imprinted gene (26). Our screen identified the second new
imprinted gene known as oxysterol-binding protein-like 1A
(OSBPL1A), encoding an oxysterol-binding protein, a family
involving in an array of metabolic processes (27). Previously, OSBPL1A has been linked
to the cellular cholesterol homeostasis as demonstrated in cases of
knock-down and overexpression of the protein. In transgenic mice
with overexpression of human OSBPL1A under scavenger receptor A
promoter, a defect of cholesterol efflux from cholesterol-loaded
macrophages to HDL particles was found (5). The role of OSBPL1A has so far not been
investigated in the intestine, the liver and tissues attributable
to bulk HDL biogenesis (28).
Nevertheless, the described functional relations with cholesterol
efflux to HDL or apoA-I reveal that OSBPL1A, which is produced in
the intestine (http://biogps.org/) and liver, is
possibly a contributor to the secretion and/or biogenesis of HDL
particles (29,30). In this study, we performed real-time
quantitative PCR and western blot analysis to further validate the
miRNA-mRNA regulatory relationship between miR-499a and osbpl1a,
and found that osbpl1a siRNA could not affect the miR-499a level,
and miR-499a enhanced miR-499a expression in a
concentration-dependent manner. Moreover, both mRNA and protein
levels of osbpl1a in cells transfected with miR-449a (30 and 60 nM)
or osbpl1a siRNA were remarkably down-regulated compared to NC
groups, furthermore the inhibitory effect of either 30 nM miR-499a
mimic or 60 nM miR-499a mimic on osbpl1a expression was similar
with that of osbpl1a siRNA, on the contrary both mRNA and protein
levels of osbpl1a were notably upregulated in miR-499a
low-expression cells by transfecting with anti-miR-449a (30 and 60
nM) compared to NC groups, and the osbpl1a mRNA and protein in 30
nM miR-499a mimic treatment group was comparable with 60 nM
miR-499a mimic treatment group.
Changes in the expression of miRNA genes are
believed to account for the pathogenesis of stroke, such as edema
formation, inflammation, oxidative damage, neuronal cell death,
diabetes mellitus, hypertension and atherosclerosis (31). miRNAs may be new biomarkers for
cardiovascular disorders such as high blood pressure, stroke,
diabetes mellitus and coronary artery disease (32–35).
In a Chinese population (391 healthy subjects and 296 ischemic
stroke patients), Liu and colleagues analyzed three SNPs and
observed that the frequency of the allele G of
hsa-mir-499/rs3746444 was significantly related to ischemic stroke.
miR-499 and miR-196a2 modulated CRP and Annexin A1, which are also
the common contributors to cerebral ischemia and related to
increased triglycerides, insulin resistance, BMI and blood pressure
(36–39). Although Liu et al reported
that miR-499 G allele was significantly associated with increased
risk of ischemic stroke in Chinese population, we found that
miR-499/rs3746444 and miR-196a2/rs11614913 were not related to
ischemic stroke in this study (40). These two SNPs have also been
extensively investigated in other human disorders, such as cancer,
coronary heart disease and congenital heart disease (41–43).
An earlier experiment revealed that the high serum level of
miR-499A>G was significantly correlated with a longer survival
of non-small cell lung cancer, and that miRNA plays a role in tumor
biology and is correlated with the development and prognosis of
cancer (44). Typical single
nucleotide polymorphisms in pre-miRNA, rs2910164 in miR-146aG>C
and rs3746444 in miR-499A>G, have been investigated in a variety
of cancers, such as colorectal cancer, cervical squamous cell
cancer, gastric cancer and breast cancer (45,46).
In this study, we collected 46 participants with atherosclerosis to
explore the effect of rs3746444 polymorphism on the expression of
miR-499a, and found that miR-499a was overexpressed in AA group
compared with AG and GG groups, and also performed one-way ANOVA
analysis, and revealed that HDL level in AA group was much lower
than AG and GG groups.
In conclusion, the findings of this study
demonstrated that rs3746444 polymorphism influenced the expression
of miR-499a, its target gene, osbpl1a, and thereby associated with
the HDL level, making it a potential factor involved in the
mechanism of atherosclerosis.
References
|
1
|
Lloyd-Jones D, Adams R, Carnethon M, De
Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund
K, et al: American Heart Association Statistics Committee and
Stroke Statistics Subcommittee: Heart disease and stroke statistics
- 2009 update: A report from the American Heart Association
Statistics Committee and Stroke Statistics Subcommittee.
Circulation. 119:e21–e181. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davidson MH: Novel nonstatin strategies to
lower low-density lipoprotein cholesterol. Curr Atheroscler Rep.
11:67–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Johansson M, Lehto M, Tanhuanpää K, Cover
TL and Olkkonen VM: The oxysterol-binding protein homologue ORP1L
interacts with Rab7 and alters functional properties of late
endocytic compartments. Mol Biol Cell. 16:5480–5492. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van der Kant R, Fish A, Janssen L, Janssen
H, Krom S, Ho N, Brummelkamp T, Carette J, Rocha N and Neefjes J:
Late endosomal transport and tethering are coupled processes
controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci.
126:3462–3474. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan D, Jauhiainen M, Hildebrand RB, van
Dijk Willems K, Van Berkel TJ, Ehnholm C, Van Eck M and Olkkonen
VM: Expression of human OSBP-related protein 1L in macrophages
enhances atherosclerotic lesion development in LDL
receptor-deficient mice. Arterioscler Thromb Vasc Biol.
27:1618–1624. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dvinge H, Git A, Gräf S, Salmon-Divon M,
Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al:
The shaping and functional consequences of the microRNA landscape
in breast cancer. Nature. 497:378–382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pillai RS: MicroRNA function: Multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Madrigal-Matute J, Rotllan N, Aranda JF
and Fernández-Hernando C: MicroRNAs and atherosclerosis. Curr
Atheroscler Rep. 15:3222013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vickers KC, Palmisano BT, Shoucri BM,
Shamburek RD and Remaley AT: MicroRNAs are transported in plasma
and delivered to recipient cells by high-density lipoproteins. Nat
Cell Biol. 13:423–433. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao W, He HW, Wang ZM, Zhao H, Lian XQ,
Wang YS, Zhu J, Yan JJ, Zhang DG, Yang ZJ, et al: Plasma levels of
lipometabolism-related miR-122 and miR-370 are increased in
patients with hyperlipidemia and associated with coronary artery
disease. Lipids Health Dis. 11:552012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karolina DS, Tavintharan S, Armugam A,
Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF and Jeyaseelan K:
Circulating miRNA profiles in patients with metabolic syndrome. J
Clin Endocrinol Metab. 97:E2271–E2276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin
Y, Wang E, Wu M and Shen SH: Aberrant allele frequencies of the
SNPs located in microRNA target sites are potentially associated
with human cancers. Nucleic Acids Res. 35:4535–4541. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeon YJ, Kim OJ, Kim SY, Oh SH, Oh D, Kim
OJ, Shin BS and Kim NK: Association of the miR-146a, miR-149,
miR-196a2, and miR-499 polymorphisms with ischemic stroke and
silent brain infarction risk. Arterioscler Thromb Vasc Biol.
33:420–430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Motazacker MM, Pirhonen J, van Capelleveen
JC, Weber-Boyvat M, Kuivenhoven JA, Shah S, Hovingh GK, Metso J, Li
S, Ikonen E, et al: A loss-of-function variant in OSBPL1A
predisposes to low plasma HDL cholesterol levels and impaired
cholesterol efflux capacity. Atherosclerosis. 249:140–147. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mallat Z, Hugel B, Ohan J, Lesèche G,
Freyssinet JM and Tedgui A: Shed membrane microparticles with
procoagulant potential in human atherosclerotic plaques: A role for
apoptosis in plaque thrombogenicity. Circulation. 99:348–353. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rautou PE, Leroyer AS, Ramkhelawon B,
Devue C, Duflaut D, Vion AC, Nalbone G, Castier Y, Leseche G,
Lehoux S, et al: Microparticles from human atherosclerotic plaques
promote endothelial ICAM-1-dependent monocyte adhesion and
transendothelial migration. Circ Res. 108:335–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu Z, Han Y, Liu J, Jiang F, Hu H, Wang Y,
Liu Q, Gong Y and Li X: MiR-135b-5p and MiR-499a-3p promote cell
proliferation and migration in atherosclerosis by directly
targeting MEF2C. Sci Rep. 5:122762015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Libby P: Inflammation in atherosclerosis.
Nature. 420:868–874. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kakimoto Y, Kamiguchi H, Ochiai E, Satoh F
and Osawa M: MicroRNA stability in postmortem FFPE tissues:
Quantitative analysis using autoptic samples from acute myocardial
infarction patients. PLoS One. 10:e01293382015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deddens JC, Colijn JM, Oerlemans MI,
Pasterkamp G, Chamuleau SA, Doevendans PA and Sluijter JP:
Circulating microRNAs as novel biomarkers for the early diagnosis
of acute coronary syndrome. J Cardiovasc Transl Res. 6:884–898.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiao J, Shen B, Li J, Lv D, Zhao Y, Wang F
and Xu J: Serum microRNA-499 and microRNA-208a as biomarkers of
acute myocardial infarction. Int J Clin Exp Med. 7:136–141.
2014.PubMed/NCBI
|
|
24
|
Olkkonen VM, Johansson M, Suchanek M, Yan
D, Hynynen R, Ehnholm C, Jauhiainen M, Thiele C and Lehto M: The
OSBP-related proteins (ORPs): Global sterol sensors for
co-ordination of cellular lipid metabolism, membrane trafficking
and signalling processes? Biochem Soc Trans. 34:389–391. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jirtle RL and Skinner MK: Environmental
epigenomics and disease susceptibility. Nat Rev Genet. 8:253–262.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okamura K, Yamada Y, Sakaki Y and Ito T:
An evolutionary scenario for genomic imprinting of Impact lying
between nonimprinted neighbors. DNA Res. 11:381–390. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jaworski CJ, Moreira E, Li A, Lee R and
Rodriguez IR: A family of 12 human genes containing
oxysterol-binding domains. Genomics. 78:185–196. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zannis VI, Fotakis P, Koukos G, Kardassis
D, Ehnholm C, Jauhiainen M and Chroni A: HDL biogenesis,
remodeling, and catabolism. Handb Exp Pharmacol. 224:53–111. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Johansson M, Bocher V, Lehto M, Chinetti
G, Kuismanen E, Ehnholm C, Staels B and Olkkonen VM: The two
variants of oxysterol binding protein-related protein-1 display
different tissue expression patterns, have different intracellular
localization, and are functionally distinct. Mol Biol Cell.
14:903–915. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Phillips MC: Molecular mechanisms of
cellular cholesterol efflux. J Biol Chem. 289:24020–24029. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tan JR, Koo YX, Kaur P, Liu F, Armugam A,
Wong PT and Jeyaseelan K: microRNAs in stroke pathogenesis. Curr
Mol Med. 11:76–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li S, Zhu J, Zhang W, Chen Y, Zhang K,
Popescu LM, Ma X, Lau WB, Rong R, Yu X, et al: Signature microRNA
expression profile of essential hypertension and its novel link to
human cytomegalovirus infection. Circulation. 124:175–184. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan KS, Armugam A, Sepramaniam S, Lim KY,
Setyowati KD, Wang CW and Jeyaseelan K: Expression profile of
MicroRNAs in young stroke patients. PLoS One. 4:e76892009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zampetaki A, Kiechl S, Drozdov I, Willeit
P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E,
et al: Plasma microRNA profiling reveals loss of endothelial
miR-126 and other microRNAs in type 2 diabetes. Circ Res.
107:810–817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fichtlscherer S, De Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T,
Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang B, Chen J, Li Y, Zhang J, Li D, Huang
Z, Cai B, Li L, Shi Y, Ying B, et al: Association of polymorphisms
in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis
in the Chinese Han population. Hum Immunol. 73:101–106. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luthra R, Singh RR, Luthra MG, Li YX,
Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, et al:
MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of
annexin A1 downregulation in cancers. Oncogene. 27:6667–6678. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Solito E, McArthur S, Christian H, Gavins
F, Buckingham JC and Gillies GE: Annexin A1 in the brain -
undiscovered roles? Trends Pharmacol Sci. 29:135–142. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wessel J, Moratorio G, Rao F, Mahata M,
Zhang L, Greene W, Rana BK, Kennedy BP, Khandrika S, Huang P, et
al: C-reactive protein, an ‘intermediate phenotype’ for
inflammation: Human twin studies reveal heritability, association
with blood pressure and the metabolic syndrome, and the influence
of common polymorphism at catecholaminergic/beta-adrenergic pathway
loci. J Hypertens. 25:329–343. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Ma Y, Zhang B, Wang SX, Wang XM and
Yu JM: Genetic polymorphisms in pre-microRNAs and risk of ischemic
stroke in a Chinese population. J Mol Neurosci. 52:473–480. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu J, Hu Z, Xu Z, Gu H, Yi L, Cao H, Chen
J, Tian T, Liang J, Lin Y, et al: Functional variant in
microRNA-196a2 contributes to the susceptibility of congenital
heart disease in a Chinese population. Hum Mutat. 30:1231–1236.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L,
Zeng Y, Miao R, Jin G, Ma H, et al: Genetic variants of miRNA
sequences and non-small cell lung cancer survival. J Clin Invest.
118:2600–2608. 2008.PubMed/NCBI
|
|
43
|
Xiong XD, Cho M, Cai XP, Cheng J, Jing X,
Cen JM, Liu X, Yang XL and Suh Y: A common variant in pre-miR-146
is associated with coronary artery disease risk and its mature
miRNA expression. Mutat Res. 761:15–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu
Y, Chen Y, Xu L, Zen K, Zhang C, et al: Serum microRNA signatures
identified in a genome-wide serum microRNA expression profiling
predict survival of non-small-cell lung cancer. J Clin Oncol.
28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang J, Bi J, Liu X, Li K, Di J and Wang
B: Hsa-miR-146a polymorphism (rs2910164) and cancer risk: A
meta-analysis of 19 case-control studies. Mol Biol Rep.
39:4571–4579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Catucci I, Yang R, Verderio P, Pizzamiglio
S, Heesen L, Hemminki K, Sutter C, Wappenschmidt B, Dick M, Arnold
N, et al: Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as
low-penetrance alleles in German and Italian familial breast cancer
cases. Hum Mutat. 31:E1052–E1057. 2010. View Article : Google Scholar : PubMed/NCBI
|