P-element-induced wimpy testis (PIWI)-interacting
RNAs (piRNAs) belong to a new class of ncRNAs that have been
associated with many cancers (1).
piRNAs are involved in the gene regulation process in which certain
nucleotides bind coding regions in gene promoters (2). piRNAs function in the epigenetic
regulation of DNA methylation (3),
transposable silencing and chromatin modification (4). PIWI is a type of Argonaute protein
that binds to piRNAs and carries out unique functions in somatic
and germ cells, and stably expressed piRNAs have also been detected
in human blood (5). piRNAs may be
used as biomarkers for cancer diagnosis (6). Four PIWI proteins have been discovered
in humans: PIWI1, PIWI2, PIWI3, and PIWI4 (7–10).
Furthermore, PIWI expression levels are associated with different
types of cancers and clinical stages (11–13).
Additionally, piRNAs have been linked to proliferation, apoptosis
(14), genomic instability
(15), invasion, and metastasis
(16) in cancer cells. The levels
of PIWI and piRNAs were revealed to be significantly altered
between tumor tissues and non-tumor tissues. The clinical
pathological features of tumors are associated with PIWI and piRNA
expression. Therefore, additional studies are needed to understand
the role of piRNAs in cancer and their epigenetic mechanisms and to
shed light on the potential of piRNAs for the diagnosis and
prediction of clinical cancer stages.
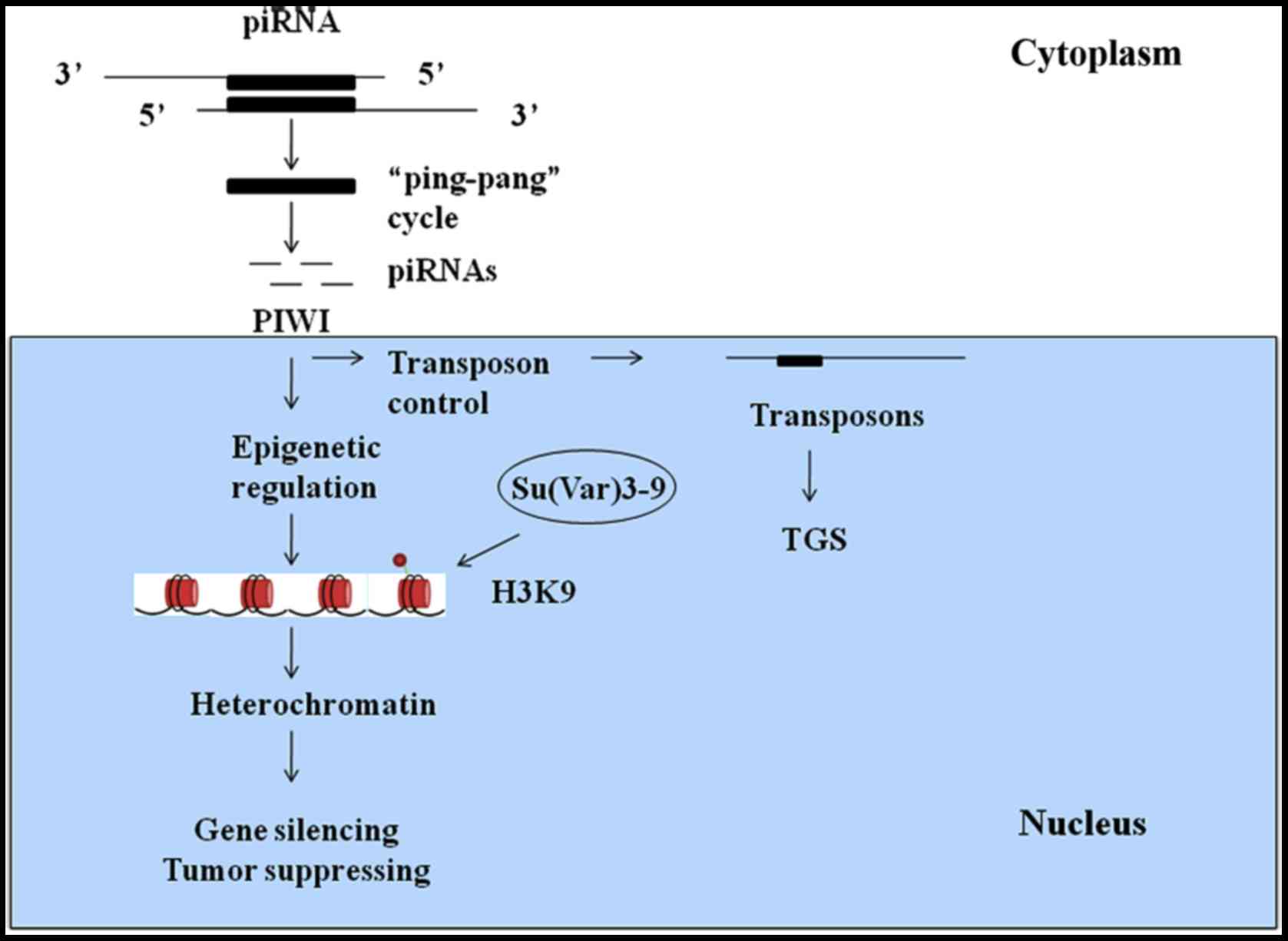
piRNAs play crucial roles in protecting genomic
stability by inhibiting transposon activity and maintaining minimum
levels of transposons in germ line, mammalian cells and other cell
types (22). piRNAs reside in
clusters within heterochromatin-euchromatin boundaries and exhibit
repeat-rich regions with ancient fragmented transposon copies.
Amplification of piRNAs occurs through the ‘ping-pang’ cycle
(23). This cycle is initiated by
the emergence of a primary piRNA from piRNA clusters. Primary
piRNAs are antisense sequences to expressed transposons and have
the ability to cleave their targets, while secondary piRNAs form
the AGO3 complex. The AGO3-bound piRNAs interact with transposon
targets, which include antisense transposon sequences. This
interaction then produces antisense piRNAs, and the ‘ping-pang’
cycle continues.
PIWI proteins have been reported to be selectively
expressed in precancerous stem cells, tumor cell lines and cells of
various malignancies (24). PIWI
and Aubergine (Aub) proteins accumulate in the pole plasm and
transfer maternal piRNAs into germ line cells (25). It was previously believed that PIWI
proteins could maintain genomic integrity in animal germ cells by
silencing transposons. Without these proteins, piRNAs cannot
inactivate transposons within a single generation. In mammals, the
PIWI proteins MILI, MIWI2 and intracisternal A-particle (IAP) act
as retrotransposons and function as transposon-inhibiting factors
via transposon gene silencing (TGS) (26). MILI is active in the cytosol, and
MIWI2 is active in the nucleus (27). MILI, also known as PIWIL2, and
MIWI2, also known as PIWIL4, suppress transposons in the cytoplasm
and nucleus. MIWI is a slicer similar to Argonaute family members,
which include PIWI proteins. The function of the slicer MIWI
depends on its binding motif, which has a conserved Argonaute
domain sequence (27). A high
degree of MIWI complementarity is required for piRNA targeting.
MIWI-associated piRNAs endonucleolytically cleave their target
RNAs. Therefore, silencing of transposons depends on the piRNAs
involved. Using next-generation sequencing, we can analyze histone
modifications and methylation across entire genomes. Moreover, we
can identify ncRNAs that participate in these regulatory processes.
This technology provides a link between epigenetic modifications
and transcription (28).
Transcriptional silencing, heterochromatin
formation, transgene silencing, HP1α alterations, histone
modifications and transposon suppression are all associated with
PIWI, piRNAs, the piRNA pathway and specific transposable elements
(29).
As mediators of eukaryotic evolution, transposable
elements are classified as either retrotransposons or DNA
transposons. Notably, retrotransposons can cause genomic variations
(30), alter chromatin structures
and change the expression of nearby genes by integrating into
genomic locations. Retrotransposons include long terminal repeat
(LTR) and non-LTR retrotransposons. LTRs are similar to
retroviruses in their structure, and non-LTRs are similar to mRNAs
(31).
LTRs can encode structural proteins to form
virus-like particles (VLPs), which can regulate gene transcription.
RNA derived from LTRs can be reverse-transcribed into cDNA and can
thereby integrate into the genome (32). RNA polymerase II located at the
5′end of LTRs can transcribe LTRs. These RNA molecules are then
packaged into viral particles and use the reverse transcription
machinery to generate full-length DNA. The first priming event
occurs between the 5′end and the 3′end of LTR, and the second
priming event occurs near the 3′end of LTR leading to production of
a double-stranded cDNA molecule through an additional strand
transfer. Thus, the cDNA is integrated into the host genome. The
main difference between retrotransposons and infectious
retroviruses is the presence of the envelope (env) gene in the
latter, which enables viruses to infect other cells. In contrast,
exon open reading frames (ORFs) are found in retrotransposons and
retroviral genomes (33).
Non-LTRs are classified as either autonomous
non-LTRs, such as LINEs, or non-autonomous LTRs, such as short
interspersed nuclear elements (SINEs) (34). Retrotransposition-competent LINE1
includes a 5′-untranslated region (UTR) that is rich in
CpG-islands; ORF1, which binds to RNA and ORF2, which encodes
proteins such as endonuclease, reverse transcriptase and
cysteine-histidine-rich domains. Similar to mammalian RNAs, LINE1
RNA has a poly (A) tail at the 3′-UTR. RNA polymerase II
transcribes ORF1 and ORF2 in the cytoplasm (35).
Mutations are generated during the process of
evolutionary change. Proofreading polymerases repair damaged DNA
sequences and eliminate potential mutations. However, some piRNAs
can also mediate the activity of transposable elements (36). HP1, a chromatin-organizing protein,
can affect transposon activity by regulating piRNA expression or by
directly mediating the expression of transposons. We can therefore
conclude that some chromatin-organizing proteins, such as HP1, act
upstream or downstream of piRNAs to regulate transposons. Mutations
in Aub, PIWI, and Su(var)205 are known to increase the activity of
transposable elements in germline cells (37,38)
(Fig. 1).
Uncontrolled transposons threaten genomic integrity,
and these alterations can be transferred to the next generation.
piRNAs bind with their partner PIWI to recognize and silence
transposable elements in germ cells. Both cytoplasmic and nuclear
PIWI proteins target the genome to mediate transcriptional
silencing. In mice, transposon inhibition by piRNAs occurs via DNA
methylation at CpG islands in the sequences of transposable
elements. During this process, the piRNA pathway mediates and
maintains high levels of the repressive H3K9me3 mark in LINE
regions in germ cells. Furthermore, piRNAs recognize full-length
elements of the actively transposing LINE family (39).
Scientists recently identified two piRNA pathway
proteins that are related to transposon silencing (40). The first, CG9754, is a downstream
piRNA pathway factor that participates in the nuclear PIWI-piRNA
complex involved in transcriptional silencing and heterochromatin
formation. CG9754 is the first protein to target RNA or DNA, in
heterochromatin and transcriptional silencing (41–43).
PIWI is only able to silence downstream proteins if it is bound to
piRNAs that are engaged with target genes (44–46).
Recruitment of CG9754 directly to DNA or indirectly to RNA results
in potent transcriptional silencing. Thus, CG9754 is sufficient to
induce transcriptional silencing by binding to RNA. In ovarian
somatic cells (OSCs), CG9754 is a downstream protein of PIWI
(47–49). When vectors were used to delete
CG9754, a decrease in the level of H3K9me3 was observed, and the
insertion of transposable elements into transcriptional genomic
regions was repressed. CG9754 mediates heterochromatin formation
and integrates other cell factors, such as HP1α, which induces
transcriptional silencing. SetDB1 is downstream of HP1α and CG9754,
an important HMT effector (50–52).
EXD1 is the second important piRNA pathway protein. MILI slicing
acts as a switch that initiates piRNA processing. Slicing activates
different types of primary piRNAs in MILI and MIWI. EXD1 is a
component of the PET (PIWI-EXD1-TDRD12) complex, which mediates
transcriptional gene silencing (53).
piRNAs exhibit a long lifetime in cells, even though
they are only 24 to 30 nt long (54), indicating that they are not easily
degraded and may exist in cell nuclei and cytoplasm for a longer
time than other RNAs (55). piRNAs
play an important role in cancer development (56). Real-time PCR and next-generation
sequencing analyses have made the relationship between piRNAs and
carcinogenesis increasingly clear (11). Compared with non-cancerous tissues,
the expression of piR-651, piR-823, piR-4987, piR-20365, piR-20485
and piR-20583 was revealed to be altered in cancer cell lines
(12,13).
piRNAs can serve as biomarkers for the prognosis,
diagnosis and clinical evaluation of cancer (8). The expression level of piR-823 in
gastric cancer tissues was revealed to be lower than that in
non-cancerous gastric tissues (57). A transfection-mediated increase in
the piR-823 level inhibited the growth of gastric cancer cells.
These results were also observed in nude mice. Thus, piR-823 may be
a potential marker for cancer diagnosis (58).
The expression levels of PIWIL2, PIWIL4, and piR-823
were associated with the tumor-node-metastasis (TNM) stage of renal
cell carcinoma (10). Additionally,
increased piR-4987 expression regulated lymph node metastasis in
breast cancer.
Here, we present an example to clarify the biomarker
characteristics of piRNAs. Through small RNA sequencing and
real-time PCR analysis of frozen benign kidney tissues and renal
cell carcinoma tissues, 26,991 piRNAs were revealed to be expressed
in kidney tissues (59). In the
tumor samples, 19 types of piRNAs were found to be deregulated,
including 2 types that were upregulated and 17 types that were
downregulated. Furthermore, differentiation was much more obvious
in the metastatic renal cell carcinoma samples (16). By comparing the localized tumor
samples to the metastatic samples, 46 piRNAs were found to be
aberrantly expressed, 44 of which were upregulated, while only 2
were downregulated. The increased piR-32051, piR-39894, and
piR-43607 are similar in length, highly homologous and derived from
the same piRNA cluster on chromosome 17. Increased expression of
these piRNAs is found in late-stage tumors, which means they are
highly associated with renal cell carcinoma metastasis (60).
Significantly increased piR-651 levels have been
observed in non-small cell lung carcinoma (NSCLC). In A549 cells,
an NSCLC cell line, piR-651 increased cell viability and
metastasis. The expression of piR-651 in A549 cells decreased the
proportion of cells arrested in the G0/G1 phase, thereby promoting
proliferation. Oncogenes and tumor suppressor genes can be
detected, and piR-651 was revealed to promote cancer growth via
cyclin D1 and CDK4. These results have also been verified in lung
cancer tissues from patients (61).
piR-1245 has been revealed to be overexpressed in
colorectal cancer, lung, breast, stomach, bladder, kidney and
prostate cancer, indicating its significant role in carcinogenesis.
Poor differentiation, advanced T stage, lymph node and distant
metastasis were revealed to be closely related to higher expression
of piR-1245 in colorectal cancer (CRC). piR-1245 also played a
crucial role in clinical pathology and revealed poor overall
survival (OS) in colorectal cancer patients. piR-1245 directly
targeted ATF3, BTG1, DUSP1, FAS, NFKBIA, UPP1, SESN2, TP53INP1 and
MDX1 to regulate tumor progression. Thus, it may be a prognostic
biomarker in CRC (62).
Similar to miRNAs, which are stable in tissues,
piRNAs are relatively stable and can be used to obtain reliable
results in quantitative piRNA expression studies (63). piRNAs can be detected in patient
plasma (64), suggesting their
potential use as biomarkers to predict TNM stage and disease
prognosis (65). Furthermore,
piRNAs can serve as a switch allowing tumors to proliferate and
metastasize (66). Additionally,
piRNAs can be indicative of patient outcomes and can be helpful in
selecting effective surgical methods, radiotherapy and chemotherapy
to prolong patient survival time (67).
PIWI proteins have been found in human cancers, such
as breast, lung, gastric, hepatocellular, colon, renal cell
carcinoma, endometrial and ovarian cancer. PIWI acts via a distinct
pathway to regulate carcinogenesis. PIWIL can affect transcription,
causing an increase in Bcl-XL, Stat3 and cyclin D1 expression
(68).
Cancer stem cells (CSCs), contain epigenetic
alterations and signaling pathways characteristic of stem cells
including self-renewal capacity, rapid proliferation and multiline
age differentiation. PIWIs may be cancer testis antigens (CATs) and
act as oncogenes or constitute markers for CSCs (69). Metastatic cancer cells appear to
undergo epithelial-mesenchymal transition (EMT) and CSC-like
phenotype (70). PIWIL2 expression
was associated with altered expression of EMT markers.
As a member of the PIWI gene family, MILI binds to
piRNAs and plays multiple roles in gene silencing (71) and chromatin remodeling (72). Transposon methylation has been
observed in tumor cell lines and many types of human cancer
(73–75). MILI was revealed to control the
activity of LINE1 by methylating its CpG island (76). The hypomethylation of LINE1
increased the risk of cancer development and may be an indicator of
cancer grade and lymph node metastasis (77,78).
Wang et al found that MILI affected melanoma cell metastasis
and cancer-related gene expression by regulating LINE1 methylation
(79). MILI is expressed in the
melanoma cell line B16 but not in the highly metastatic mouse
melanoma model B16BL6. Notably, knockdown of MILI in B16 cells
activated MAGEA expression and increased cell migration, whereas
MILI overexpression in the B16BL6 model inhibited MAGEA expression
and decreased cell migration, yielding the opposite results
(65,80). Depletion of MILI/MIWI2 in mice led
to reduced DNA methylation of LTR-retrotransposon promoter regions
(81). Thus, LINE1 methylation by
MILI was revealed to controls the expression of cancer-related
genes and cell migration, and MILI plays a key role in melanoma
metastasis and tumor progression.
The function of piRNAs in tumors is related to
transposable elements and changes in chromatin structures that are
caused by epigenetic alterations, such as DNA hypomethylation
(60). For example, in HeLa cells,
piRNAs play an important role in inhibiting transposons by
interacting with the HILI protein (82). The relationship between piRNAs and
epigenetics is elaborated above, and the epigenetic changes account
for a large proportion in tumors. Perhaps piRNAs and transposable
elements, upstream or downstream of epigenetic alterations in
tumors, affect the metastasis ability of tumors. Next, we will
illustrate the epigenetic mechanisms of cancer metastasis related
to piRNAs.
Epigenetic evaluations involve examination of DNA
methylation profiles, certain RNA expression profiles (87) and histone modification profiles
(88). Epigenetic alterations do
not involve changes in DNA sequence (89). DNA methylation and histone
modification are the major types of epigenetic alteration (90). In normal tissues, DNA methylation
can prevent X chromosome activation and gene mutations (91), and histone modifications can
dynamically regulate gene activity.
Here, mechanisms related to epigenetic alterations
are briefly summarized. First, DNA methylation not only affects the
expression of individual genes but also affects DNA domains by
interacting with nucleosomes, thus altering DNA packaging. DNA
methylation is inevitable and occurs on the cytosine of CpG
dinucleotides. In mammalian cells, methylation is conferred by four
main DNA methyltransferases (DNMTs): DNMT1 (92), DNMT3A (93), DNMT3B (94,95)
and DNMT3L (96). DNMT1 adds methyl
groups to hemi-methylated CpG sites, DNMT3A and DNMT3B methylate
novel CpG sites, and DNMT3L interacts with DNMT3A and 3B to
facilitate methylation of retrotransposons. DNA demethylation and
remethylation comprise a balanced process that is disrupted in
cancer cells (97). Tumor
progression and metastasis occur due to changes in DNA methylation.
Studies investigating DNA methylation in promoter regions have been
fruitful, leading to the discovery of adenomatous polyposis coli
(APC), retinoic acid receptor β-2 and H-cadherin (98), which are also associated with tumor
progression (99). In primary
testicular tumors, scientists detected a gain of 5′end promoter CpG
island methylation of the PIWIL1, PIWIL2, PIWIL4 and TDRD1 genes in
association with transcriptional silencing. The DNMT3L/PIWIL2/TDRD1
complex is responsible for the loss of DNA methylation at LINE1 and
IA transposons (100). The extent
of DNA methylation in tumor tissues is lower than that in normal
tissues, and the degree of DNA hypomethylation increases with the
progression of malignancy. DNA hypomethylation is conducive to
mitotic recombination, leading to chromosomal deletions and
translocations, which promote chromosomal rearrangements (101). DNA methylation in malignant cells
can reactivate genomic DNA repeat elements, such as long
interspersed element 1 (LINE1) (77) and Alu (102). These demethylated transposons can
be transcribed or transposed to other genomic regions and disrupt
the genome (103). Transposable
elements, which are abundant in the human genome, are highly
mutagenic due to their ability to target protein-coding genes for
insertion, resulting in chromosome breakage and promoting
illegitimate genome rearrangement.
Another mechanism related to cancer is histone
modification. There are two types of nuclear chromatin, namely,
heterochromatin and euchromatin (104). In normal tissues, heterochromatin
is stable during various cell cycle phases and silences during
transcription (105), and the
genes in euchromatin are actively transcribed. As part of an
interplay with DNA methylation, facultative heterochromatin, which
is associated with allelic exclusion, genomic imprinting, X
chromosome stabilization (106),
immunoglobulin (Igh/Igk) and T-cell receptor-α and -β (107) gene loci, is vital for normal cell
lineage development and cell differentiation via somatic
methylation and inactivation of germline-specific genes (108).
Initiation, propagation and maintenance of
heterochromatin are largely controlled by trimethylation of lysine
9 on histone H3 (H3K9me3) and other synergistic epigenetic
modifications (109). Chromosomal
regions that are abundant in repetitive DNA help H3K9me3 stabilize
constitutive heterochromatin, facultative heterochromatin and
intermediate or transient heterochromatin, the 3 heterochromatin
subtypes. By preventing abnormal chromosome segregation,
recombination and DNA replication, H3K9me3 regulates constitutive
heterochromatin to stabilize genomic integrity (110).
Histone H3 determines the formation of different
chromatin structures. Methylation of the N-terminal lysine of
histone H3 is vital for well-documented histone modifications.
H3K9me3, H3K36me3, and possibly H3K79me3 facilitate the opening of
the chromatin configuration to form euchromatin, which is also
associated with serine 10 phosphorylation and lysine 9 acetylation
of histone H3 for active transcription of genes. H3K9me3 and
H3K27me3 mainly function in the initiation, propagation and
maintenance of highly compact heterochromatin to silence gene
expression (111).
Histone proteins expose DNA euchromatin to
facilitate the binding of transcription factors. Methylation and
acetylation are the two major mechanisms by which histone function
is regulated (112).
Methyltransferases and demethylases modify the lysines of histone
H3 to form mono-, di-, or tri-methylated lysines, which contribute
to chromatin structure and gene transcription (113). Histone methyltransferases (HMTs)
regulate histone proteins by transferring methyl groups from
S-adenosylmethionine to lysine or arginine residues in histones.
Histone acetylation is regulated by histone acetyl-transferases
(HATs) and deacetylases (HDACs) (114). Not surprisingly, there are other
histone modifications. The balance of these modifications and their
effects on histone structure ultimately coordinate DNA exposure.
Histone methylation is a key event in gene transcription, and it is
plausible to speculate that this type of modification can regulate
DNA replication, recombination, and damage repair (115).
H3K9me3 recruits transcriptional repressors such as
repressor element 1 silencing transcription factor (REST) and
CoREST, which contain histone deacetylases (HDACs) (116,117), H3K4me3 demethylases LSD1 (118) and Rbp4 (119), to actively transcribe gene loci,
leading to gene blocking and suppression of gene transcription
(120). Recruitment of DNMTs as
well as additional histone methylases is responsible for localized
chromatin condensation when the tethering of HP1α (heterochromatin
protein) and HP1ß to H3K9me2 or H3K9me3 triggers gene silencing
(121). Thus, H3K9me3 acts as a
natural brake to prevent unnecessary over-transcription of actively
expressed genes. Attenuation of H3K9me3 by either
over-transcription of demethylases or deficiency of H3K9
methyltransferases will therefore lead to sustained expression of
the genes involved in either cell cycle transition or proliferation
(122).
Changes in chromatin structure that are caused by
epigenetic alterations can contribute to cancer development. In
experimental studies using mice lacking SUV39, a methyltransferase
that acts on H3K9, enhanced genomic instability and incidence of
B-cell lymphoma were observed (123,124). In addition, polymorphisms of SUV39
can increase lung cancer risk due to piRNA instability and
decreased levels of H3K9me3. H3K9 methyltransferases include G9a
for mono- and di-methylation and SUV39h1/h2 for di- and
tri-methylation of H3K9. Similarly, low levels of RIZI (125), another methyltransferase of H3K9,
are frequently observed in lung cancer, breast cancer,
hepatocellular carcinoma, colon cancer, neuroblastoma, and melanoma
(126). Methyltransferases, such
as SUV39 and RZZI act as tumor suppressors, while some demethylases
may have oncogenic activity. The low expression of these
methyltransferases in tumor cells may be the result of increased
cell proliferation, apoptosis resistance and poor differentiation
(127). Global regulation of
H3K9me3 has been observed in several human cancers, including
colorectal, ovarian, and lung cancer, all of which are
characterized by deficiency or elevated activity of H3K9
methyltransferases or changes in the expression of H3K9
demethylases (128,129).
Epigenetic changes associated with piRNA expression,
affect genes associated with malignant phenotypes (VE-cadherin,
VEGF-C, PAX8, Keratin 7, CD13, laminin, urokinase, α3-integrin
subunit and c-met) (8).
Detection of some forms of methylation in tumors can
be diagnostic of cancer. 5-Hydroxymethylcytosine (5hmC) in cfDNA,
which is found in blood originating from different tissues, is the
basis of noninvasive prenatal diagnostic tests, organ transplant
rejection diagnostics, and cancer detection. 5hmC is useful in gene
regulation and cancer pathogenesis and can be used diagnostically
to identify cancer types and track tumor stage. Li et al
observed a progressive global loss of 5hmC in cfDNA in lung cancer,
whereas disease-specific changes in the cell-free hydroxymethylome
have been observed in hepatocellular carcinoma and pancreatic
cancer (130).
DNMT3Ab plays crucial role in directing
EMT-associated metastasis in gastric cancer (GC). Increased DNMT3A
expression was revealed to be closely related to a poor survival
rate in GC, breast, lung and liver cancer. Furthermore, TNM stage
and lymph node metastasis of GC cells were more closely associated
with DNMT3A than with DNMT1 and DNMT3B. Increased expression of
DNMT3Ab was demonstrated to promote GC cell migration and invasion
as well as EMT progression. DNMT3Ab mediated the E-cadherin gene
via DNA hypermethylation and histone modifications of H3K9me2 and
H3K27me3. DNMT3Ab effectively regulated the expression of
E-cadherin via DNA hypermethylation and histone modifications of
H3K9me2 and H3K27me3. DNMT3Ab in cooperation with H3K9me2 and
H3K27me3 contributed to the transcriptional regulation of
E-cadherin in a Snail-dependent manner and multiple
metastasis-associated genes and oncogenic signaling pathways are
regulated by DNMT3Ab overexpression. Thus, it was revealed that
DNMT3Ab acts as a crucial regulator of metastasis-related genes in
GC (137) (Table I).
Gliomas with histone H3 lysine27-to-methionine
mutations primarily occur in the central nervous system of young
children, which means that there is a link between genetics and
cellular context in tumorigenesis. Through single-cell RNA
sequencing of 3321 cells from six primary H3K27M-glioma and matched
models, Filbin et al found that H3K27M-gliomas contained
cells that resembled oligodendrocyte precursor cells (OPC-like
cells). OPC-like cells tend to exhibit higher proliferation and a
greater tumor-propagating potential and some of these cells display
PDGFRA signaling (138).
miRNAs, short interfering RNAs (siRNAs) and piRNAs
are involved in the regulation of mRNA transcripts,
chromatin-mediated gene silencing, and DNA rearrangement. miRNAs
accelerate de-adenylation of the poly(A) tail and downregulate the
expression of some pathways, thereby downregulating the expression
of hundreds of target genes. siRNAs can also control transposons.
RDR2-dependent siRNAs, which are endo-siRNAs, silence transposons,
retroelements and DNA methylation (142). siRNAs bind to a nascent RNA being
transcribed at their target site, resulting in RNA-induced
transcriptional silencing (RITS) and formation of the RNA-directed
RNA polymerase complex (RDRC) at the site of intended
heterochromatin formation, which in turn results in TGS. TGS occurs
in the nucleus. During this process, siRNAs guide miRNAs to modify
chromatin, which also influences the cell cycle (143). miR-127 and miR-136 are released
near 2 CpG islands in the Rtl1 transcript and thus regulate
RISC-mediated cleavage of the maternal transcript, resulting in
late-fetal or neonatal lethality (144).
In mice, the miR-290-295 miRNA cluster was revealed
to act as a transcriptional repressor of the DNMTs, Dnmt3a and
Dnmt3b, resulting in the appearance of long telomeres and increased
telomere recombination. The expression of this cluster remained
high in undifferentiated ES cells, but decreased after ES cell
differentiation. This example indicates a direct or indirect
function of miRNAs in regulating genes involved in self-renewal or
differentiation by affecting methylation.
piRNA expression detected in both patient lymph
nodes and serum samples is related to tumor treatment failure.
piRNAs can act as tumor promoters or cancer suppressors and can
participate in other carcinoma cell activities. piRNAs and PIWI can
serve as biomarkers for the prognosis, diagnosis and clinical
evaluation of cancer, and can be helpful in selecting effective
surgical methods, radiotherapy and chemotherapy to prolong patient
survival time. piRNA can serve as a switch, allowing tumors to
proliferate and metastasize.
In mammals, PIWI proteins function as
transposon-inhibiting factors through TGS. PIWI acts via a distinct
pathway to regulate carcinogenesis by affecting transcription and
the expression of other carcinoma-related genes that reduce
apoptosis and increase cell proliferation and transformation.
Transcriptional silencing, heterochromatin formation, transgene
silencing, HP1α alteration, histone modifications and transposon
suppression are all associated with PIWI, piRNAs and the piRNA
pathway. Transposable elements are classified as either
retrotransposons or DNA transposons. Retrotransposons can be
further divided into LTR and non-LTR groups. Non-LTRs are divided
into the autonomous non-LTRs LINEs and non-autonomous LTRs SINEs.
Chromatin-organizing proteins such as HP1, Aub and Su(var)205, act
upstream or downstream of piRNAs to regulate transposons.
Hypomethylation of LINE1 increases the risk of cancer development
and may be an indicator of cancer grade and lymph node metastasis.
LINE1 methylation by MILI was revealed to control the expression of
cancer-related genes and cell migration, and MILI played a key role
in melanoma metastasis and tumor progression. In the sequences of
transposable elements, the piRNA pathway mediates and maintains
high levels of the repressive H3K9me3 mark in LINE1 regions in germ
cells. There are two identified pathway proteins that are related
to transposon silencing: CG9754 and EXD1.
Perhaps piRNAs, PIWI, transposable elements and
piRNA pathways, upstream or downstream of the epigenetic
alterations in tumors, affect the metastasis ability of tumors. In
addition, epigenetic alteration of piRNAs, cancer stem cells, CpG
island methylation and EMT all participate in cancer
metastasis.
Epigenetic alterations associated with cancer
metastasis involve DNA methylation, histone modification and
certain RNA expression profiles. First, DNA methylation affects the
expression of individual genes and DNA domains. The degree of DNA
hypomethylation increases as tumors progress and metastasize. DNA
methylation and histone modification can activate genomic DNA
repeat elements, such as LINE1 and Alu, which can be transcribed or
transposed to other genomic regions and disrupt the genome,
resulting in chromosome breakage and illegitimate genome
rearrangement.
H3K9me3 and other synergistic epigenetic
modifications control heterochromatin, but in tumors, that balance
has been disrupted. Therefore, with the ability to stabilize
genomic integrity by preventing abnormal chromosome segregation,
recombination and DNA replication, H3K9me3 and H3K27me3 mainly
function in the initiation, propagation and maintenance of highly
compact heterochromatin to silence gene expression. Methylation and
acetylation are the two major mechanisms that regulate histone
function. HMTs regulate histone proteins. Histone acetylation is
regulated by HATs and HDACs. Several HMTs exhibit tumor suppressor
functions, and some demethylases exhibit oncogenic activity.
H3K9me3 recruits transcriptional repressors such as REST and
CoREST. The low expression of methyltransferases such as SUV39 and
RZZI in tumor cells may be the result of increased cell
proliferation and apoptosis resistance and poor differentiation.
Global regulation of H3K9me3 has been observed in several human
cancers, including colorectal, ovarian and lung cancer, all of
which are characterized by deficiency or elevated activity of H3K9
methyltransferases or altered expression of H3K9 demethylases. 5hmC
can be used to identify cancer type and track tumor stage. DNMT3Ab
cooperated with H3K9me2 and H3K27me3 played a crucial role in
directing EMT-associated metastasis in gastric cancer.
Understanding the epigenetic mechanisms of tumor
metastasis related to piRNAs can assist in the identification of
new tumor markers and treatments. piRNAs can be indicative of
patient outcomes and can be helpful in selecting effective surgical
methods, radiotherapy and chemotherapy to prolong patient survival
time. piRNAs can be used as biomarkers to predict TNM stage and
disease prognosis. Since preventing tumor metastasis is still a
formidable problem, studies on the potential of piRNAs and
epigenetic alternations for diagnosis and prediction of clinical
cancer stages are greatly needed.
Not applicable.
The present review was supported by the grant
2017Y001 from the First Affiliated Hospital of Harbin Medical
University Research Innovation Fund. This review was supported by
the First Affiliated Hospital of Harbin Medical University and the
Laboratory of Hepatosplenic Surgery Center of Heilongjiang
Province. The authors are solely responsible for the content, and
this manuscript does not represent the official views of the First
Affiliated Hospital of Harbin Medical University.
Not applicable.
BC, SZ and JL conceived and designed the study. JL
wrote the manuscript. SZ prepared the figure and the table. BC
wrote the ncRNA part, reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H,
Seitz H, Horwich MD, Syrzycka M, Honda BM, et al: Collapse of
germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs
in flies. Cell. 137:509–521. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shrey K, Suchit A, Nishant M and Vibha R:
RNA interference: Emerging diagnostics and therapeutics tool.
Biochem Biophys Res Commun. 386:273–277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Collins LJ and Penny D: The RNA
infrastructure: Dark matter of the eukaryotic cell? Trends Genet.
25:120–128. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Malone CD and Hannon GJ: Small RNAs as
guardians of the genome. Cell. 136:656–668. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malone CD, Brennecke J, Dus M, Stark A,
McCombie WR, Sachidanandam R and Hannon GJ: Specialized piRNA
pathways act in germline and somatic tissues of the Drosophila
ovary. Cell. 137:522–535. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Freedman JE, Gerstein M, Mick E, Rozowsky
J, Levy D, Kitchen R, Das S, Shah R, Danielson K, Beaulieu L, et
al: Diverse human extracellular RNAs are widely detected in human
plasma. Nat Commun. 7:111062016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iliev R, Stanik M, Fedorko M, Poprach A,
Vychytilova-Faltejskova P, Slaba K, Svoboda M, Fabian P, Pacik D,
Dolezel J, et al: Decreased expression levels of PIWIL1, PIWIL2,
and PIWIL4 are associated with worse survival in renal cell
carcinoma patients. OncoTargets Ther. 9:217–222. 2016.
|
|
8
|
Lee JH, Schutte D, Wulf G, Füzesi L,
Radzun HJ, Schweyer S, Engel W and Nayernia K: Stem-cell protein
Piwil2 is widely expressed in tumors and inhibits apoptosis through
activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 15:201–211.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taubert H, Greither T, Kaushal D, Würl P,
Bache M, Bartel F, Kehlen A, Lautenschläger C, Harris L, Kraemer K,
et al: Expression of the stem cell self-renewal gene Hiwi and risk
of tumour-related death in patients with soft-tissue sarcoma.
Oncogene. 26:1098–1100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Liu Y, Shen X, Zhang X, Chen X,
Yang C and Gao H: The PIWI protein acts as a predictive marker for
human gastric cancer. Int J Clin Exp Pathol. 5:315–325.
2012.PubMed/NCBI
|
|
11
|
Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z,
Zhou H and Li QN: piRNA, the new non-coding RNA, is aberrantly
expressed in human cancer cells. Clin Chim Acta. 412:1621–1625.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang G, Hu H, Xue X, Shen S, Gao E, Guo
G, Shen X and Zhang X: Altered expression of piRNAs and their
relation with clinicopathologic features of breast cancer. Clin
Transl Oncol. 15:563–568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Law PT, Qin H, Ching AK, Lai KP, Co NN, He
M, Lung RW, Chan AW, Chan TF and Wong N: Deep sequencing of small
RNA transcriptome reveals novel non-coding RNAs in hepatocellular
carcinoma. J Hepatol. 58:1165–1173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang
L, Chen L, Chu ZB, Tang B, Wang K, et al: piRNA-823 contributes to
tumorigenesis by regulating de novo DNA methylation and
angiogenesis in multiple myeloma. Leukemia. 29:196–206. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luteijn MJ and Ketting RF:
PIWI-interacting RNAs: from generation to transgenerational
epigenetics. Nat Rev Genet. 14:523–534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Wu X, Gao H, Jin JM, Li AX, Kim YS,
Pal SK, Nelson RA, Lau CM, Guo C, et al: Piwi-Interacting RNAs
(piRNAs) are dysregulated in renal cell carcinoma and associated
with tumor metastasis and cancer-specific survival. Mol Med.
21:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gigek CO, Chen ES, Calcagno DQ, Wisnieski
F, Burbano RR and Smith MA: Epigenetic mechanisms in gastric
cancer. Epigenomics. 4:279–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Potter VR: Initiation and promotion in
cancer formation: the importance of studies on intercellular
communication. Yale J Biol Med. 53:367–384. 1980.PubMed/NCBI
|
|
19
|
Lujambio A, Calin GA, Villanueva A, Ropero
S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso
MS, Faller WJ, et al: A microRNA DNA methylation signature for
human cancer metastasis. Proc Natl Acad Sci USA. 105:13556–13561.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tiwari VK, McGarvey KM, Licchesi JD, Ohm
JE, Herman JG, Schübeler D and Baylin SB: PcG proteins, DNA
methylation, and gene repression by chromatin looping. PLoS Biol.
6:2911–2927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nowacka-Zawisza M and Wisnik E: DNA
methylation and histone modifications as epigenetic regulation in
prostate cancer (Review). Oncol Rep. 38:2587–2596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weick EM and Miska EA: piRNAs: From
biogenesis to function. Development. 141:3458–3471. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han BW and Zamore PD: piRNAs. Curr Biol.
24:R730–R733. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aravin AA, Sachidanandam R, Girard A,
Fejes-Toth K and Hannon GJ: Developmentally regulated piRNA
clusters implicate MILI in transposon control. Science.
316:744–747. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ross RJ, Weiner MM and Lin H: PIWI
proteins and PIWI-interacting RNAs in the soma. Nature.
505:353–359. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Houwing S, Kamminga LM, Berezikov E,
Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz
E, Moens CB, et al: A role for Piwi and piRNAs in germ cell
maintenance and transposon silencing in Zebrafish. Cell. 129:69–82.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aravin AA, Sachidanandam R, Bourc'his D,
Schaefer C, Pezic D, Toth KF, Bestor T and Hannon GJ: A piRNA
pathway primed by individual transposons is linked to de novo DNA
methylation in mice. Mol Cell. 31:785–799. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ghildiyal M and Zamore PD: Small silencing
RNAs: an expanding universe. Nat Rev Genet. 10:94–108. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peng JC and Lin H: Beyond transposons: The
epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr
Opin Cell Biol. 25:190–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feschotte C and Pritham EJ: DNA
transposons and the evolution of eukaryotic genomes. Annu Rev
Genet. 41:331–368. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kubo S, Seleme MC, Soifer HS, Perez JL,
Moran JV, Kazazian HH Jr and Kasahara N: L1 retrotransposition in
nondividing and primary human somatic cells. Proc Natl Acad Sci
USA. 103:8036–8041. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pelisson A, Mejlumian L, Robert V, Terzian
C and Bucheton A: Drosophila germline invasion by the endogenous
retrovirus gypsy: Involvement of the viral env gene. Insect Biochem
Mol Biol. 32:1249–1256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leblanc P, Desset S, Giorgi F, Taddei AR,
Fausto AM, Mazzini M, Dastugue B and Vaury C: Life cycle of an
endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol.
74:10658–10669. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brouha B, Schustak J, Badge RM,
Lutz-Prigge S, Farley AH, Moran JV and Kazazian HH Jr: Hot L1s
account for the bulk of retrotransposition in the human population.
Proc Natl Acad Sci USA. 100:5280–5285. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Alisch RS, Garcia-Perez JL, Muotri AR,
Gage FH and Moran JV: Unconventional translation of mammalian
LINE-1 retrotransposons. Genes Dev. 20:210–224. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Baer CF, Miyamoto MM and Denver DR:
Mutation rate variation in multicellular eukaryotes: causes and
consequences. Na Rev Genet. 8:619–631. 2007. View Article : Google Scholar
|
|
37
|
Belinco C, Diprima SN, Wolff RE, Thorp MW,
Buschette JT and Simmons MJ: Cytotype regulation in Drosophila
melanogaster: synergism between telomeric and non-telomeric P
elements. Genet Res. 91:383–394. 2009. View Article : Google Scholar
|
|
38
|
Simmons MJ, Peterson MP, Thorp MW,
Buschette JT, DiPrima SN, Harter CL and Skolnick MJ: piRNA-mediated
transposon regulation and the germ-line mutation rate in Drosophila
melanogaster males. Mutat Res. 773:16–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pezic D, Manakov SA, Sachidanandam R and
Aravin AA: piRNA pathway targets active LINE1 elements to establish
the repressive H3K9me3 mark in germ cells. Genes Dev. 28:1410–1428.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Czech B, Preall JB, McGinn J and Hannon
GJ: A transcriptome-wide RNAi screen in the Drosophila ovary
reveals factors of the germline piRNA pathway. Mol Cell.
50:749–761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Handler D, Meixner K, Pizka M, Lauss K,
Schmied C, Gruber FS and Brennecke J: The genetic makeup of the
Drosophila piRNA pathway. Mol Cell. 50:762–777. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Muerdter F, Guzzardo PM, Gillis J, Luo Y,
Yu Y, Chen C, Fekete R and Hannon GJ: A genome-wide RNAi screen
draws a genetic framework for transposon control and primary piRNA
biogenesis in Drosophila. Mol Cell. 50:736–748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sarot E, Payen-Groschene G, Bucheton A and
Pelisson A: Evidence for a piwi-dependent RNA silencing of the
gypsy endogenous retrovirus by the Drosophila melanogaster flamenco
gene. Genetics. 166:1313–1321. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Le Thomas A, Rogers AK, Webster A, Marinov
GK, Liao SE, Perkins EM, Hur JK, Aravin AA and Tóth KF: Piwi
induces piRNA-guided transcriptional silencing and establishment of
a repressive chromatin state. Genes Dev. 27:390–399. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sienski G, Batki J, Senti KA, Dönertas D,
Tirian L, Meixner K and Brennecke J: Silencio/CG9754 connects the
Piwi-piRNA complex to the cellular heterochromatin machinery. Genes
Dev. 29:2258–2271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sienski G, Donertas D and Brennecke J:
Transcriptional silencing of transposons by Piwi and maelstrom and
its impact on chromatin state and gene expression. Cell.
151:964–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gunawardane LS, Saito K, Nishida KM,
Miyoshi K, Kawamura Y, Nagami T, Siomi H and Siomi MC: A
slicer-mediated mechanism for repeat-associated siRNA 5′end
formation in Drosophila. Science. 315:1587–1590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Niki Y, Yamaguchi T and Mahowald AP:
Establishment of stable cell lines of Drosophila germ-line stem
cells. Proc Natl Acad Sci USA. 103:16325–16330. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Saito K, Ishizu H, Komai M, Kotani H,
Kawamura Y, Nishida KM, Siomi H and Siomi MC: Roles for the Yb body
components Armitage and Yb in primary piRNA biogenesis in
Drosophila. Genes Dev. 24:2493–2498. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ayyanathan K, Lechner MS, Bell P, Maul GG,
Schultz DC, Yamada Y, Tanaka K, Torigoe K and Rauscher FJ III:
Regulated recruitment of HP1 to a euchromatic gene induces
mitotically heritable, epigenetic gene silencing: A mammalian cell
culture model of gene variegation. Genes Dev. 17:1855–1869. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li Y, Danzer JR, Alvarez P, Belmont AS and
Wallrath LL: Effects of tethering HP1 to euchromatic regions of the
Drosophila genome. Development. 130:1817–1824. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rangan P, Malone CD, Navarro C, Newbold
SP, Hayes PS, Sachidanandam R, Hannon GJ and Lehmann R: piRNA
production requires heterochromatin formation in Drosophila. Curr
Biol. 21:1373–1379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang Z, Chen KM, Pandey RR, Homolka D,
Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA
and Pillai RS: PIWI slicing and EXD1 drive biogenesis of nuclear
piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell.
61:138–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He W, Wang Z, Wang Q, Fan Q, Shou C, Wang
J, Giercksky KE, Nesland JM and Suo Z: Expression of HIWI in human
esophageal squamous cell carcinoma is significantly associated with
poorer prognosis. BMC Cancer. 9:4262009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
He G, Chen L, Ye Y, Xiao Y, Hua K,
Jarjoura D, Nakano T, Barsky SH, Shen R and Gao JX: Piwil2
expressed in various stages of cervical neoplasia is a potential
complementary marker for p16INK4a. Am J Transl Res. 2:156–169.
2010.PubMed/NCBI
|
|
56
|
Rajasethupathy P, Antonov I, Sheridan R,
Frey S, Sander C, Tuschl T and Kandel ER: A role for neuronal
piRNAs in the epigenetic control of memory-related synaptic
plasticity. Cell. 149:693–707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cui L, Lou Y, Zhang X, Zhou H, Deng H,
Song H, Yu X, Xiao B, Wang W and Guo J: Detection of circulating
tumor cells in peripheral blood from patients with gastric cancer
using piRNAs as markers. Clin Biochem. 44:1050–1057. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cheng J, Deng H, Xiao B, Zhou H, Zhou F,
Shen Z and Guo J: piR-823, a novel non-coding small RNA,
demonstrates in vitro and in vivo tumor suppressive activity in
human gastric cancer cells. Cancer Lett. 315:12–17. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Siomi MC, Sato K, Pezic D and Aravin AA:
PIWI-interacting small RNAs: The vanguard of genome defence. Nat
Rev Mol Cell Biol. 12:246–258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li D, Luo Y, Gao Y and Yang Y, Wang Y, Xu
Y, Tan S, Zhang Y, Duan J and Yang Y: piR-651 promotes tumor
formation in non-small cell lung carcinoma through the upregulation
of cyclin D1 and CDK4. Int J Mol Med. 38:927–936. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Weng W, Liu N, Toiyama Y, Kusunoki M,
Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y and Goel A: Novel
evidence for a PIWI-interacting RNA (piRNA) as an oncogenic
mediator of disease progression, and a potential prognostic
biomarker in colorectal cancer. Mol Cancer. 17:162018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zeng Y, Qu LK, Meng L, Liu CY, Dong B,
Xing XF, Wu J and Shou CC: HIWI expression profile in cancer cells
and its prognostic value for patients with colorectal cancer. Chin
Med J. 124:2144–2149. 2011.PubMed/NCBI
|
|
65
|
Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K,
Jarjoura D, Nakano T, Ramesh GK, Shapiro CL, et al: Piwil2 is
expressed in various stages of breast cancers and has the potential
to be used as a novel biomarker. Int J Clin Exp Pathol. 3:328–337.
2010.PubMed/NCBI
|
|
66
|
Grochola LF, Greither T, Taubert H, Möller
P, Knippschild U, Udelnow A, Henne-Bruns D and Würl P: The stem
cell-associated Hiwi gene in human adenocarcinoma of the pancreas:
expression and risk of tumour-related death. Br J Cancer.
99:1083–1088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Mei Y, Clark D and Mao L: Novel dimensions
of piRNAs in cancer. Cancer Lett. 336:46–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tan Y, Liu L, Liao M, Zhang C, Hu S, Zou
M, Gu M and Li X: Emerging roles for PIWI proteins in cancer. Acta
Biochim Biophys Sin. 47:315–324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Litwin M, Szczepanska-Buda A, Piotrowska
A, Dziegiel P and Witkiewicz W: The meaning of PIWI proteins in
cancer development. Oncol Lett. 13:3354–3362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tamura S, Isobe T, Ariyama H, Nakano M,
Kikushige Y, Takaishi S, Kusaba H, Takenaka K, Ueki T, Nakamura M,
et al: Ecadherin regulates proliferation of colorectal cancer stem
cells through NANOG. Oncol Rep. 40:693–703. 2018.PubMed/NCBI
|
|
71
|
Liu J, Carmell MA, Rivas FV, Marsden CG,
Thomson JM, Song JJ, Hammond SM, Joshua-Tor L and Hannon GJ:
Argonaute2 is the catalytic engine of mammalian RNAi. Science.
305:1437–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yin H and Lin H: An epigenetic activation
role of Piwi and a Piwi-associated piRNA in Drosophila
melanogaster. Nature. 450:304–308. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen Y, Hu W, Lu Y, Jiang S, Li C, Chen J,
Tao D, Liu Y, Yang Y and Ma Y: A TALEN-based specific transcript
knock-down of PIWIL2 suppresses cell growth in HepG2 tumor cell.
Cell Pprolif. 47:448–456. 2014. View Article : Google Scholar
|
|
74
|
Lee JH, Jung C, Javadian-Elyaderani P,
Schweyer S, Schütte D, Shoukier M, Karimi-Busheri F, Weinfeld M,
Rasouli-Nia A, Hengstler JG, et al: Pathways of proliferation and
antiapoptosis driven in breast cancer stem cells by stem cell
protein piwil2. Cancer Res. 70:4569–4579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang H, Ren Y, Xu H, Pang D, Duan C and
Liu C: The expression of stem cell protein Piwil2 and piR-932 in
breast cancer. Surg Oncol. 22:217–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lee E, Iskow R, Yang L, Gokcumen O,
Haseley P, Luquette LJ III, Lohr JG, Harris CC, Ding L, Wilson RK,
et al: Landscape of somatic retrotransposition in human cancers.
Science. 337:967–971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Andreotti G, Karami S, Pfeiffer RM,
Hurwitz L, Liao LM, Weinstein SJ, Albanes D, Virtamo J, Silverman
DT, Rothman N and Moore LE: LINE1 methylation levels associated
with increased bladder cancer risk in pre-diagnostic blood DNA
among US (PLCO) and European (ATBC) cohort study participants.
Epigenetics. 9:404–415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Daskalos A, Nikolaidis G, Xinarianos G,
Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field
JK and Liloglou T: Hypomethylation of retrotransposable elements
correlates with genomic instability in non-small cell lung cancer.
Int J Cancer. 124:81–87. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang X, Jiang C, Fu B, Zhu R, Diao F, Xu
N, Chen Z, Tao W and Li CJ: MILI, a PIWI family protein, inhibits
melanoma cell migration through methylation of LINE1. Biochem
Biophys Res Commun. 457:514–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ye Y, Yin DT, Chen L, Zhou Q, Shen R, He
G, Yan Q, Tong Z, Issekutz AC, Shapiro CL, et al: Identification of
Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One.
5:e134062010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kuramochi-Miyagawa S, Watanabe T, Gotoh K,
Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri
TW, et al: DNA methylation of retrotransposon genes is regulated by
Piwi family members MILI and MIWI2 in murine fetal testes. Genes
Dev. 22:908–917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lu Y, Li C, Zhang K, Sun H, Tao D, Liu Y,
Zhang S and Ma Y: Identification of piRNAs in Hela cells by massive
parallel sequencing. BMB Rep. 43:635–641. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Virani S, Colacino JA, Kim JH and Rozek
LS: Cancer epigenetics: A brief review. ILAR J. 53:359–369. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chik F, Szyf M and Rabbani SA: Role of
epigenetics in cancer initiation and progression. Adv Exp Med Biol.
720:91–104. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cavalli LR, Urban CA, Dai D, de Assis S,
Tavares DC, Rone JD, Bleggi-Torres LF, Lima RS, Cavalli IJ, Issa
JP, et al: Genetic and epigenetic alterations in sentinel lymph
nodes metastatic lesions compared to their corresponding primary
breast tumors. Cancer Genet Cytogenet. 146:33–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hinoue T, Weisenberger DJ, Lange CP, Shen
H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk
CM, et al: Genome-scale analysis of aberrant DNA methylation in
colorectal cancer. Genome Res. 22:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Azad N, Zahnow CA, Rudin CM and Baylin SB:
The future of epigenetic therapy in solid tumours-lessons from the
past. Nat Rev Clin Oncol. 10:256–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jones PA and Taylor SM: Cellular
differentiation, cytidine analogs and DNA methylation. Cell.
20:85–93. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Merry CR, Forrest ME, Sabers JN, Beard L,
Gao XH, Hatzoglou M, Jackson MW, Wang Z, Markowitz SD and Khalil
AM: DNMT1-associated long non-coding RNAs regulate global gene
expression and DNA methylation in colon cancer. Hum Mol Genet.
24:6240–6253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liu T, Wu X, Chen T, Luo Z and Hu X:
Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem
cell-like phenotypes through repressing Wnt/β-catenin signaling.
Clin Cancer Res. 24:1748–1760. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang LH, Huang J, Wu CR, Huang LY, Cui J,
Xing ZZ and Zhao CY: Downregulation of miR29b targets DNMT3b to
suppress cellular apoptosis and enhance proliferation in pancreatic
cancer. Mol Med Rep. 17:2113–2120. 2018.PubMed/NCBI
|
|
95
|
Yang L, Hou J, Cui XH, Suo LN and Lv YW:
RG108 induces the apoptosis of endometrial cancer Ishikawa cell
lines by inhibiting the expression of DNMT3B and demethylation of
HMLH1. Eur Rev Med Pharmacol Sci. 21:5056–5064. 2017.PubMed/NCBI
|
|
96
|
Heo J, Lim J, Lee S, Jeong J, Kang H, Kim
Y, Kang JW, Yu HY, Jeong EM, Kim K, et al: Sirt1 regulates DNA
methylation and differentiation potential of embryonic stem cells
by antagonizing Dnmt3l. Cell Rep. 18:1930–1945. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Guo Y, Wang M, Jia X, Zhu H, Zhi Y and
Yuan L: Wnt signaling pathway upregulates DNMT1 to trigger NHERF1
promoter hypermethylation in colon cancer. Oncol Rep. 40:1165–1173.
2018.PubMed/NCBI
|
|
98
|
Zochbauer-Muller S, Gazdar AF and Minna
JD: Molecular pathogenesis of lung cancer. Ann Rev Physiol.
64:681–708. 2002. View Article : Google Scholar
|
|
99
|
Nakajima NI, Niimi A, Isono M, Oike T,
Sato H, Nakano T and Shibata A: Inhibition of the HDAC/Suv39/G9a
pathway restores the expression of DNA damage-dependent major
histocompatibility complex class I-related chain A and B in cancer
cells. Oncol Rep. 38:693–702. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ferreira HJ, Heyn H, del Muro Garcia X,
Vidal A, Larriba S, Muñoz C, Villanueva A and Esteller M:
Epigenetic loss of the PIWI/piRNA machinery in human testicular
tumorigenesis. Epigenetics. 9:113–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sciamanna I, Vitullo P, Curatolo A and
Spadafora C: A reverse transcriptase-dependent mechanism is
essential for murine preimplantation development. Genes. 2:360–373.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Akers SN, Moysich K, Zhang W, Lai Collamat
G, Miller A, Lele S, Odunsi K and Karpf AR: LINE1 and Alu
repetitive element DNA methylation in tumors and white blood cells
from epithelial ovarian cancer patients. Gynecol Oncol.
132:462–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Slotkin RK and Martienssen R: Transposable
elements and the epigenetic regulation of the genome. Nat Rev
Genet. 8:272–285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Shilatifard A: Chromatin modifications by
methylation and ubiquitination: implications in the regulation of
gene expression. Annu Rev Biochem. 75:243–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Grewal SI and Jia S: Heterochromatin
revisited. Nat Rev Genet. 8:35–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Heard E: Delving into the diversity of
facultative heterochromatin: the epigenetics of the inactive X
chromosome. Curr Opin Genet Dev. 15:482–489. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Corcoran AE: Immunoglobulin locus
silencing and allelic exclusion. Semin Immunol. 17:141–154. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Skok JA, Gisler R, Novatchkova M, Farmer
D, de Laat W and Busslinger M: Reversible contraction by looping of
the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol.
8:378–387. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
109
|
Klose RJ and Zhang Y: Regulation of
histone methylation by demethylimination and demethylation. Nat Rev
Mol Cell Biol. 8:307–318. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Feldman N, Gerson A, Fang J, Li E, Zhang
Y, Shinkai Y, Cedar H and Bergman Y: G9a-mediated irreversible
epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat
Cell Biol. 8:188–194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Lee E, Wang J, Jung Y, Cackowski FC and
Taichman RS: Reduction of two histone marks, H3k9me3 and H3k27me3
by epidrug induces neuroendocrine differentiation in prostate
cancer. J Cell Biochem. 119:3697–3705. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ooi L and Wood IC: Chromatin crosstalk in
development and disease: lessons from REST. Nat Rev Genet.
8:544–554. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Shi Y: Histone lysine demethylases:
emerging roles in development, physiology and disease. Nat Rev
Genet. 8:829–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yang M, Gocke CB, Luo X, Borek D, Tomchick
DR, Machius M, Otwinowski Z and Yu H: Structural basis for
CoREST-dependent demethylation of nucleosomes by the human LSD1
histone demethylase. Mol Cell. 23:377–387. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Grewal SI and Elgin SC: Transcription and
RNA interference in the formation of heterochromatin. Nature.
447:399–406. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Lee J, Ko J and Yi JY: Histone deacetylase
inhibitor (HDACi) upregulates activin A and activates the Smad
signaling pathway in melanomas. J Dermatol Sci. 90:13–20. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Terranova-Barberio M, Thomas S and Munster
PN: Host histone acetylation unlocks HDAC inhibitor potential.
Oncotarget. 8:106161–106162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Liu C, Liu L, Chen X, Cheng J, Zhang H,
Zhang C, Shan J, Shen J and Qian C: LSD1 stimulates
cancer-associated fibroblasts to drive Notch3-dependent
self-renewal of liver cancer stem-like cells. Cancer Res.
78:938–949. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Fei W, Chen L, Chen J, Shi Q, Zhang L, Liu
S, Li L, Zheng L and Hu X: RBP4 and THBS2 are serum biomarkers for
diagnosis of colorectal cancer. Oncotarget. 8:92254–92264. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Smith CD, Shu S, Mungall CJ and Karpen GH:
The Release 5.1 annotation of Drosophila melanogaster
heterochromatin. Science. 316:1586–1591. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Smallwood A, Esteve PO, Pradhan S and
Carey M: Functional cooperation between HP1 and DNMT1 mediates gene
silencing. Genes Dev. 21:1169–1178. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Peters AH, O'Carroll D, Scherthan H,
Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner
M, Kohlmaier A, et al: Loss of the Suv39h histone
methyltransferases impairs mammalian heterochromatin and genome
stability. Cell. 107:323–337. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Yoon KA, Hwangbo B, Kim IJ, Park S, Kim
HS, Kee HJ, Lee JE, Jang YK, Park JG and Lee JS: Novel
polymorphisms in the SUV39H2 histone methyltransferase and the risk
of lung cancer. Carcinogenesis. 27:2217–2222. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhao Z, Hu Y, Shen X, Lao Y, Zhang L, Qiu
X, Hu J, Gong P, Cui H, Lu S, et al: HBx represses RIZ1 expression
by DNA methyltransferase 1 involvement in decreased miR-152 in
hepatocellular carcinoma. Oncol Rep. 37:2811–2818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Gibbons RJ: Histone modifying and
chromatin remodelling enzymes in cancer and dysplastic syndromes.
Hum Mol Genet. 14:R85–R92. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lakshmikuttyamma A, Takahashi N, Pastural
E, Torlakovic E, Amin HM, Garcia-Manero G, Voralia M, Czader M,
DeCoteau JF and Geyer CR: RIZ1 is potential CML tumor suppressor
that is down-regulated during disease progression. J Hematol Oncol.
2:282009. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Espino PS, Drobic B, Dunn KL and Davie JR:
Histone modifications as a platform for cancer therapy. J Cell
Biochem. 94:1088–1102. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Hamamoto R, Furukawa Y, Morita M, Iimura
Y, Silva FP, Li M, Yagyu R and Nakamura Y: SMYD3 encodes a histone
methyltransferase involved in the proliferation of cancer cells.
Nat Cell Biol. 6:731–740. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Li W, Zhang X, Lu X, You L, Song Y, Luo Z,
Zhang J, Nie J, Zheng W, Xu D, et al: 5-Hydroxymethylcytosine
signatures in circulating cell-free DNA as diagnostic biomarkers
for human cancers. Cell Res. 27:1243–1257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Pulukuri SM, Estes N, Patel J and Rao JS:
Demethylation-linked activation of urokinase plasminogen activator
is involved in progression of prostate cancer. Cancer Res.
67:930–939. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Stepanova V, Dergilev KV, Holman KR,
Parfyonova YV, Tsokolaeva ZI, Teter M, Atochina-Vasserman EN,
Volgina A, Zaitsev SV, Lewis SP, et al: Urokinase-type plasminogen
activator (uPA) is critical for progression of tuberous sclerosis
complex 2 (TSC2)-deficient tumors. J Biol Chem. 292:20528–20543.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Foekens JA, Peters HA, Look MP, Portengen
H, Schmitt M, Kramer MD, Brünner N, Jänicke F, Meijer-van Gelder
ME, Henzen-Logmans SC, et al: The urokinase system of plasminogen
activation and prognosis in 2780 breast cancer patients. Cancer
Res. 60:636–643. 2000.PubMed/NCBI
|
|
134
|
Miyake H, Hara I, Yamanaka K, Gohji K,
Arakawa S and Kamidono S: Elevation of serum levels of
urokinase-type plasminogen activator and its receptor is associated
with disease progression and prognosis in patients with prostate
cancer. Prostate. 39:123–129. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhang Z, Wang J, Wang X, Song W, Shi Y and
Zhang L: MicroRNA-21 promotes proliferation, migration, and
invasion of cervical cancer through targeting TIMP3. Arch Gynecol
Obstet. 297:433–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Qi JH, Ebrahem Q, Moore N, Murphy G,
Claesson-Welsh L, Bond M, Baker A and Anand-Apte B: A novel
function for tissue inhibitor of metalloproteinases-3 (TIMP3):
inhibition of angiogenesis by blockage of VEGF binding to VEGF
receptor-2. Nat Med. 9:407–415. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
137
|
Cui H, Hu Y, Guo D, Zhang A, Gu Y, Zhang
S, Zhao C, Gong P, Shen X, Li Y, et al: DNA methyltransferase 3A
isoform b contributes to repressing E-cadherin through cooperation
of DNA methylation and H3K27/H3K9 methylation in EMT-related
metastasis of gastric cancer. Oncogene. 37:4358–4371. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Filbin MG, Tirosh I, Hovestadt V, Shaw ML,
Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM,
et al: Developmental and oncogenic programs in H3K27M gliomas
dissected by single-cell RNA-seq. Science. 360:331–335. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Gregory PA, Bracken CP, Bert AG and
Goodall GJ: MicroRNAs as regulators of epithelial-mesenchymal
transition. Cell Cycle. 7:3112–3118. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Suarez Y and Sessa WC: MicroRNAs as novel
regulators of angiogenesis. Circ Res. 104:442–454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liang Z, Wu H, Reddy S, Zhu A, Wang S,
Blevins D, Yoon Y, Zhang Y and Shim H: Blockade of invasion and
metastasis of breast cancer cells via targeting CXCR4 with an
artificial microRNA. Biochem Biophys Res Commun. 363:542–546. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Henderson IR and Jacobsen SE: Epigenetic
inheritance in plants. Nature. 447:418–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wassenegger M, Heimes S, Riedel L and
Sanger HL: RNA-directed de novo methylation of genomic sequences in
plants. Cell. 76:567–576. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Seitz H, Youngson N, Lin SP, Dalbert S,
Paulsen M, Bachellerie JP, Ferguson-Smith AC and Cavaillé J:
Imprinted microRNA genes transcribed antisense to a reciprocally
imprinted retrotransposon-like gene. Nat Genet. 34:261–262. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Lin SP, Coan P, da Rocha ST, Seitz H,
Cavaille J, Teng PW, Takada S and Ferguson-Smith AC: Differential
regulation of imprinting in the murine embryo and placenta by the
Dlk1-Dio3 imprinting control region. Development. 134:417–426.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Sinkkonen L, Hugenschmidt T, Berninger P,
Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P and
Filipowicz W: MicroRNAs control de novo DNA methylation through
regulation of transcriptional repressors in mouse embryonic stem
cells. Nat Struct Mol Biol. 15:259–267. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Benetti R, Gonzalo S, Jaco I, Muñoz P,
Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T and Klatt P:
A mammalian microRNA cluster controls DNA methylation and telomere
recombination via Rbl2-dependent regulation of DNA
methyltransferases. Nat Struct Mol Biol. 15:268–279. 2008.
View Article : Google Scholar : PubMed/NCBI
|