Introduction
Following heart disease, cancer is the leading cause
of mortality associated with incommunicable diseases worldwide
(1). Local infiltration and distant
metastasis are major challenges faced by oncologists, ultimately
affecting the call for effective therapy (2). Due to these challenges, the correct
diagnosis of the tissue from which the cancer originated and
further prognosis for treatment remain an area of great interest.
There are several theories that have emerged to explain the
mechanisms of a poor prognosis.
A cancerous tumor is an extremely complex ecosystem
that consists of both malignant and normal cells. The latter
include the reactive tumor stroma, which contributes to the diverse
phenotypic heterogeneity of solid tumors (3). All heterogenous cancer cells are
derived from cancer stem cells (CSCs), a relatively rare subgroup
of tumor cells possessing self-renewal properties, tumorigenicity
and multilineage differentiation ability (4–8). CSCs
are considered the drivers of malignant growth, treatment
resistance, minimal residual disease and the formation of
metastases (9–11). Moreover, CSCs have been shown to
regulate tumor growth and progression (12–15).
The tumor microenvironment plays an important role
in tumor growth and metastasis formation (16–18).
It is a dynamic structure consisting of multiple components,
including interstitial tissue and extracellular matrix, both of
which influence tumor cell properties (19). Exosomes, key players in the tumor
microenvironment, are small (between 30 and 100 nm in diameter),
membrane-enclosed vesicles secreted by cells and are distributed in
bodily fluids (20,21). Transporting a variety of
biologically active substances, such as proteins and nucleic acids,
exosomes mediate local and systemic cell communication (22,23).
Accumulating evidence demonstrates that cancer-derived exosomes
play a critical role in tumor metastasis (24–26).
However, little is known about CSC-derived exosomes.
For a number of years, fibroblasts (FBs) have been
considered an important participant in a variety of aspects
involving tumor development and progression. It has been shown that
normal FBs can be induced by surrounding tumor cells to
differentiate into tumor-promoting cancer-associated fibroblasts
(CAFs) (27,28). It has been demonstrated that the
Piwi family protein, Piwil2, regulates the self-renewal of germline
stem cells and plays an important regulatory role in tumorigenesis
(29). Piwil2 is expressed in the
human testis and various tumor cells, including colon cancer
(30), breast cancer (31) and cervical cancer (32). It has been reported that the
expression of Piwil2 in malignant tumors may be related to the
incidence, development and prognosis of tumors (33–35).
In a previous study by the authors, a Piwil2 GFP lentiviral vector
was used to transduce fibroblasts, and the successfully constructed
Piwil2-induced tumor stem cells (Piwil2-iCSC) characterized by
self-renewal, multiple differentiation capabilities, cell atypia
and tumorigenic properties (36).
In this study, to elucidate the role of exosomes derived from CSCs
in tumorigenesis and cancer progression, these induced tumor stem
cells were used to generate Piwil2-iCSC-Exo and investigate their
role in the proliferation, migration and invasion of FBs.
Materials and methods
Cell lines and cell culture
Piwil2-iCSCs and FBs were obtained from the
Chongqing Key Laboratory of Child Urogenital Development and Tissue
Engineering (36). The Piwil2-iCSCs
were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with penicillin/streptomycin and 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific) depleted of bovine
exosomes by overnight ultracentrifugation at 100,000 × g, followed
by filtration through a 0.2-µm filter (Millipore). FBs were
cultured in DMEM/F-12 medium supplemented with 10% FBS. All the
cells were cultured at 37°C in a humidified 5% CO2
atmosphere. All experiments were carried out at a cell confluence
of 80 to 90%.
Exosome isolation
Exosomes were isolated through the differential
centrifugation of conditioned medium collected from Piwil2-iCSC
cultures after achieving 90% confluency. Briefly, the conditioned
medium was first subjected to serial centrifugation to remove cells
(500 × g, 10 min) and cellular debris (2,000 × g, 20 min). Later
the supernatant was filtered (0.22 µm) to remove the remaining
debris and larger vesicles and subjected to centrifugation at
100,000 × g for 70 min to pellet the exosomes. Finally, the pellets
were washed with phosphate-buffered saline (PBS) and the
ultracentrifugation protocol was repeated. The final exosome pellet
was resuspended in PBS and frozen in aliquots maintained at
−80°C.
Transmission electron microscopy
(TEM)
The presence of purified exosomes was confirmed with
transmission electron microscopy (TEM). Exosomal proteins were
measured using a bicinchoninic acid protein assay kit (BCA,
Beyotime). In brief, 10 µg exosomes were placed on a copper grid,
followed by negative staining with 2% uranyl acetate for 2 min at
room temperature. Grids were rinsed in deionized water and allowed
to dry overnight. The samples were then visualized using a Hitachi
S-3000 N electron microscope.
Nanoparticle tracking analysis
(NTA)
The size and the concentration of the exosome
particles resuspended in PBS were determined using NTA (Zetasizer
Nano ZS90, Malvern Panalytical). The diluted exosomes were injected
into the LM10 unit (Malvern Panalytical). NTA software, v2.3
(Malvern Panalytical) was used to collect and analyze the
videos.
Exosome uptake assay
The FBs (3×104 cells/well) were
inoculated into 24-well plates (Corning Inc.) pre-loaded with cell
slides and allowed to adhere overnight. Subsequently, PKH26
(MINI26; Sigma-Aldrich) pre-stained Piwil2-iCSCs exosomes according
to the manufacturer's protocol were added to the wells. After 0, 3,
6, 12 and 24, the cells were fixed in 4% paraformaldehyde for 20
min, washed 3 times with PBS, and stained with DAPI for 1 h at room
temperature. After being sealed with an anti-fluorescence quencher,
the slides were imaged using a fluorescent microscope (Nikon).
Western blot analysis
Protein samples of exosomes and cells were lysed in
RIPA buffer (Beyotime) supplemented with proteinase inhibitors
(Beyotime). The protein concentration was determined using a
bicinchoninic acid (BCA) assay kit (Beyotime). Equal amounts of
proteins (10 µg) were separated on an 8% SDS-PAGE (sodium dodecyl
sulfate-polyacrylamide gel electrophoresis) gel and transferred
onto polyvinylidene fluoride (PVDF; Millipore) membranes. The
membranes were then blocked in 5% (w/v) non-fat milk and incubated
with the primary antibodies overnight at 4°C. The following
antibodies were used: CD9 (1:1,000, ab92726; Abcam), CD63 (1:1,000,
ab134045; Abcam), heat shock protein (HSP)70 (1:1,000, ab181606;
Abcam), Piwil2 (1:1,000, ab181340; Abcam), matrix metalloproteinase
(MMP)9 (1:1,000 ab76003; Abcam), MMP2 (1:1,000, GTX104577;
Genetex), α-smooth muscle actin (α-SMA; 1:1,000, ab32575; Abcam),
Vimentin (1:1,000, GTX100619; Genetex), fibroblast-activating
protein (FAP; 1:1,000, ab207178; Abcam), GAPDH (1:1,000, ab181602;
Abcam). Finally, the membranes were incubated with HRP-conjugated
secondary antibody (1:1,000, ZB-2301, ZB-2305; Zhongshan) for 2 h
at room temperature. The immunoblots were visualized using the
Immobilon Western Chemiluminescent HRP Substrate, and the bands
were quantified, relative to GAPDH, by densitometric analysis
(GeneGnome, Syngene UK).
Optimization experiments
In the optimization phase, a dose-escalation
experiment was performed with increasing concentrations of exosomes
(50, 80, 100, 160, 200 and 320 µg/ml) using the Cell Counting Kit-8
(CCK-8, Dojindo Laboratories) following the manufacturer's protocol
to determine the optimal exosome concentration. In brief, the cells
were seeded in 96-well plates (Corning, Inc.; 3,000 cells/well) in
triplicate and cultured in DMEM/F-12 medium supplemented with 10%
FBS overnight at 37°C. The exosome suspension was then added, and
following incubation at 37°C for 24 h, the cells were treated with
the CCK-8 reagent and the absorbance was measured using a
spectrophotometer (BioTek Epoch, BioTek Instruments, Inc.) at 450
nm.
Exosome treatment
The FBs which were collected during their
logarithmic growth phase, were inoculated into the wells and were
divided into the control group (untreated), PBS-treated group (PBS)
and the exosome-treated group (Exo). As the exosomes were suspended
in PBS, the PBS-treated group served as a control for the
experiment. The concentration of the exosomes is relatively low and
the addition of exosome suspension reduces the concentration of FBS
into the culture medium. In order to control for the effect of PBS
altering the dilution of FBS on cell growth, a blank group was
added (termed the control group) to ensure that the addition of PBS
had no obvious effect on cell growth. Each experimental group was
evaluated with 5 duplicate wells and at least 3 independent
experiments were performed.
Cell proliferation assay
Cell proliferation assays were performed using a
CCK-8 assay (Dojindo Laboratories) following the manufacturer's
protocol as described above. In brief, the cells were plated in
96-well plates (Corning, Inc.; 3,000 cells/well) in triplicate and
cultured in growth medium overnight. The cells were then divided in
accordance with their experimental groups. Subsequently, the cells
were treated with the CCK-8 reagent and the absorbance was measured
by spectrophotometer (BioTek Epoch, BioTek Instruments, Inc.) at
450 nm. Proliferation rates were determined after 0, 24, 48 and 72
h of exosome treatment.
Cell migration assay
To assess the cell migratory ability, a wound
healing assay was performed. The cells were seeded at
2×105 cells/well in 6-well plates (Corning, Inc.) to
achieve a confluent monolayer. The monolayer was then scratched
with a 200 µl pipette tip to simulate a wound. After washing with
PBS, the cells were incubated in the presence or absence of the
exosomes (1% FBS; 1% FBS was used as the FB condition deteriorated
progressively after 24 h in FBS-free medium). Microscopic
evaluation was performed by a fluorescence microscope (K10587;
Nikon) after 0, 12 and 24 h of culture.
Cell invasion assay
Cell invasion ability was analyzed in a Transwell
chamber assay. First, the cells were incubated at 37°C for 24 h in
the presence or absence of exosomes in growth medium. The cells
(2×104 cells/well) were then seeded in the upper chamber
with serum-free medium, coated with 30 mg/cm2 Matrigel
(Sigma-Aldrich) and the lower layer was supplemented with 600 µl of
DMEM/F-12 medium containing 10% FBS. Following incubation at 37°C,
5% CO2 for 24 h, the cells remaining on the top surface
of the membrane (non-invasive cells) were scraped with cotton
swabs, while the cells localized on the bottom sides of the
membrane (invasive cells) were washed with PBS, fixed with cold
methanol for 20 min, stained with eosin at room temperature for 5
min and mounted. The cells migrating through the membrane were
counted in 5 randomly selected fields under a microscope (K10587;
Nikon) at magnification, ×100.
Statistical analysis
Each experiment was repeated at least 3 times, and
SPSS v24.0 software was used for all statistical analyses.
Comparisons between groups were analyzed using one-way ANOVA. In
this study, the variance of all experiments was homogeneous; thus,
the LSD post hoc test was selected for analysis. Data are expressed
as the means ± SEM. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characterization of
Piwil2-iCSC-Exo
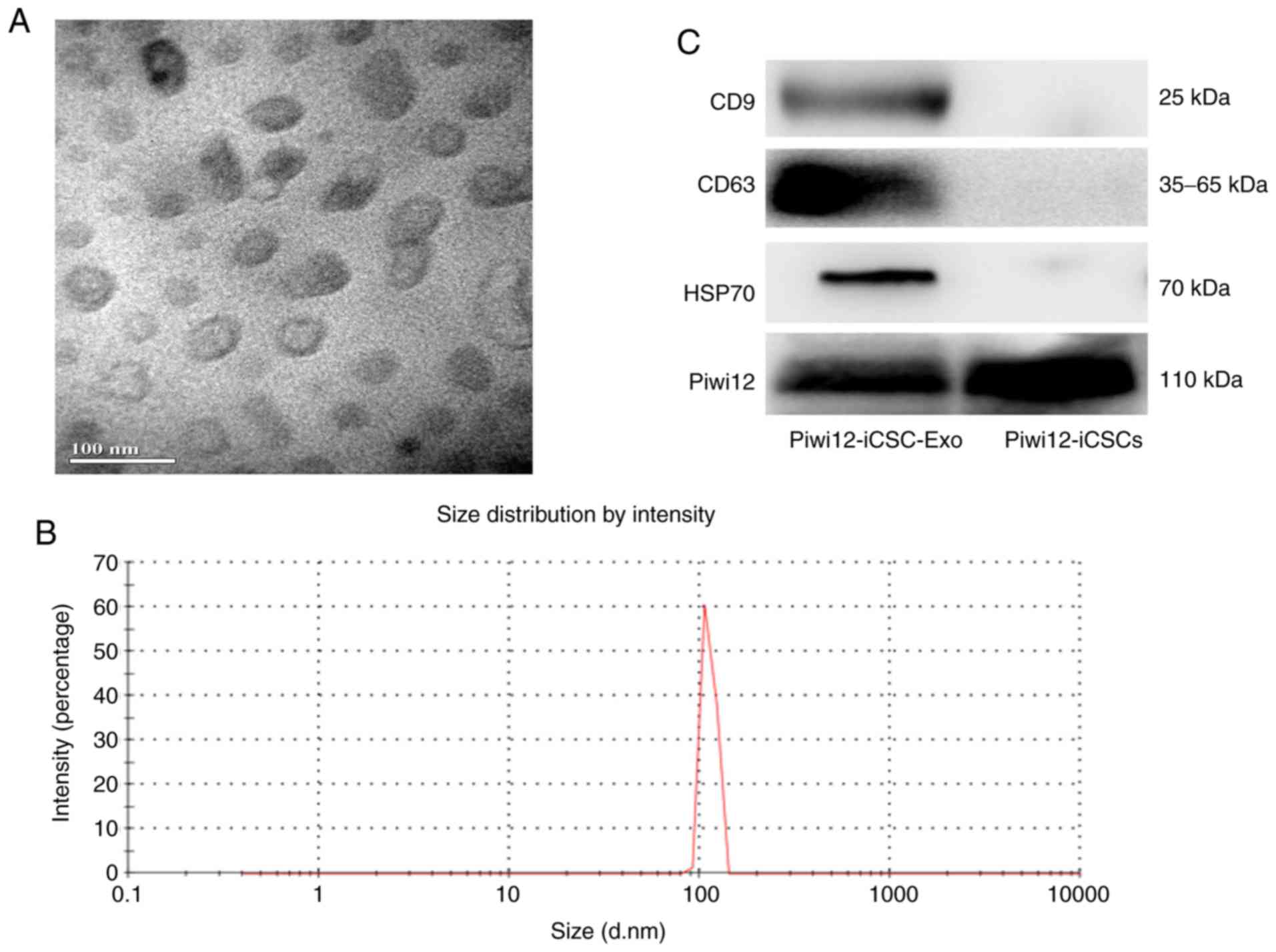
Exosome morphology was examined by TEM. As shown in
Fig. 1A, Piwil2-iCSC-Exo are lipid
bilayer-coated membrane structures ranging from 30 to 100 nm in
diameter. NTA verified a vesicle population with a maximum size of
100 nm (Fig. 1B). The difference in
the size distribution of exosomes between NTA and electron
microscopy may be due to the possible aggregation of exosomes
during the NTA analysis (37).
Further investigation of the exosome markers revealed by western
blot analysis, confirmed that Piwil2-iCSC-Exo expressed CD63, CD9,
HSP70 and Piwil2 (Fig. 1C). At
present, the majority of researchers do not detect any housekeeping
gene. In this study, GAPDH was selected as a housekeeping gene;
however, no bands were detected in the exosomes. Thus, the loading
control was not included in Fig.
1C.
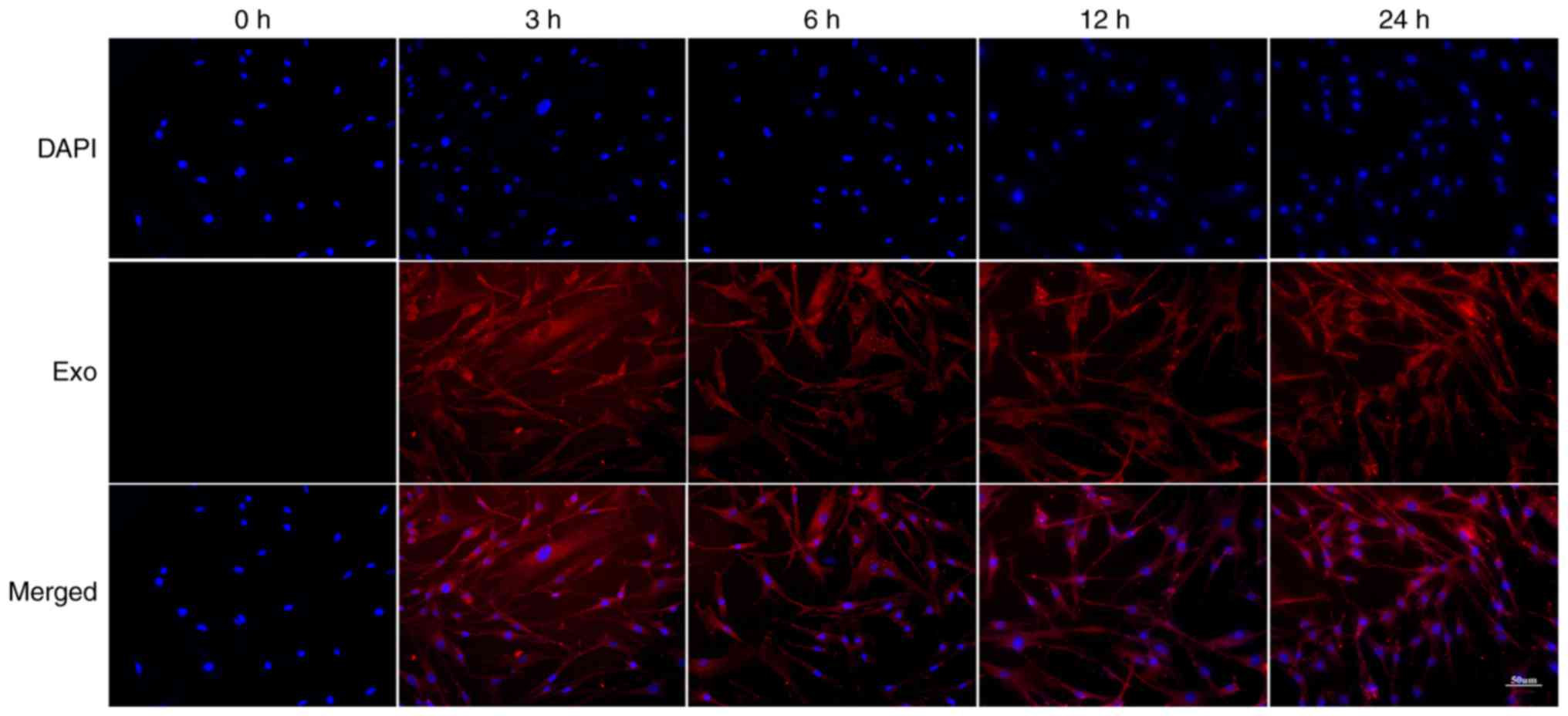
Uptake of Piwil2-iCSC-Exo by FBs
To examine whether FBs can internalize
Piwil2-iCSC-Exo, the FBs were incubated with PKH26-labeled
Piwil2-iCSC-Exo for 0, 3, 6, 12 and 24 h. The labeled
Piwil2-iCSC-Exo was shown to enter the cells in a time-dependent
manner. Following 3 and 6 h of co-culture, the exosomes entered the
cells, and at 12 h, the exosomes began to accumulate in the
cytoplasm. The accumulation of exosomes was time-dependent and
reached a maximum in this study at 24 h (Fig. 2).
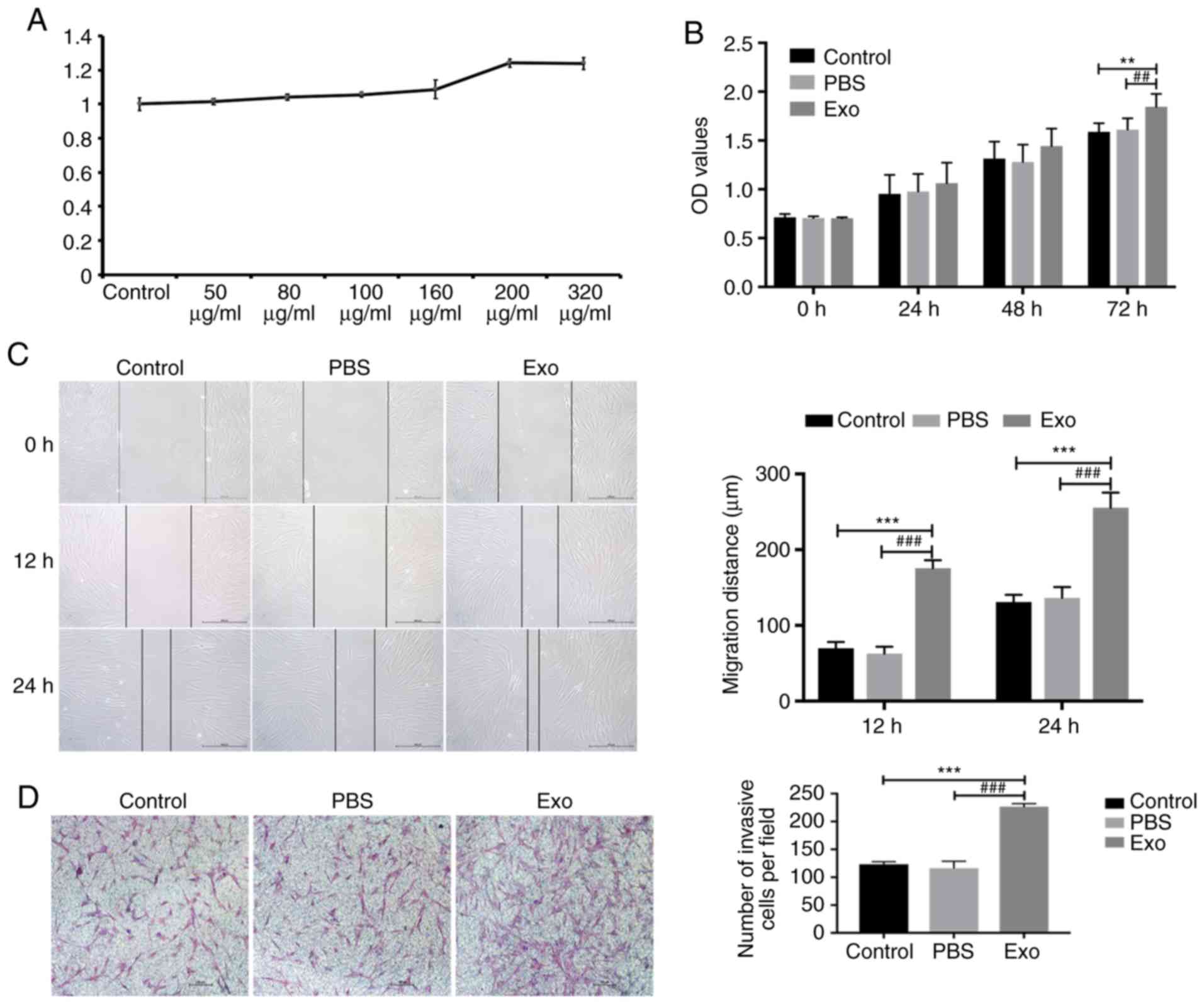
Piwil2-iCSC-Exo enhance the
proliferation, migration and invasion of FBs
In the optimization phase, it was found that the
high concentration group, namely concentrations of 200 and 320
µg/ml, markedly influenced cell proliferation (Fig. 3A). In these settings, the
concentration of 200 µg/ml was more effective than that of 320
µg/ml. The lack of dose-dependency in this case may be due to the
low concentration of exosomes in the suspension, resulting in the
significant dilution of the culture medium with the vehicle (PBS)
that in consequence, affected cell growth. Therefore, in the
subsequent experiments, the concentration of 200 µg/ml of exosomes
was used only. The potential effect of Piwil2-iCSC-Exo on the
proliferation of FBs was analyzed by CCK-8 cell proliferation
assay. Compared to the control groups, the exosomes exhibited an
increased cell proliferation rate at 72 h (Fig. 3B). The continued exploration into
the effects of Piwil2-iCSC-Exo on FB migration was carried out by
performing a wound healing assay. Following 12 h of incubation with
the exosomes, FB motility significantly increased and the wound was
almost closed after 24 h (Fig. 3C).
No significant differences were observed between the control group
and PBS group at 12 and 24 h (P>0.05; Fig. 3B). Furthermore, in order to
investigate whether Piwil2-iCSC-Exo affect the invasion of FBs, a
Transwell assay was performed. Similar to the wound healing assay,
the results revealed that the exosomes enhanced the invasive
ability of the FBs (Fig. 3D)
compared with that observed in the control groups.
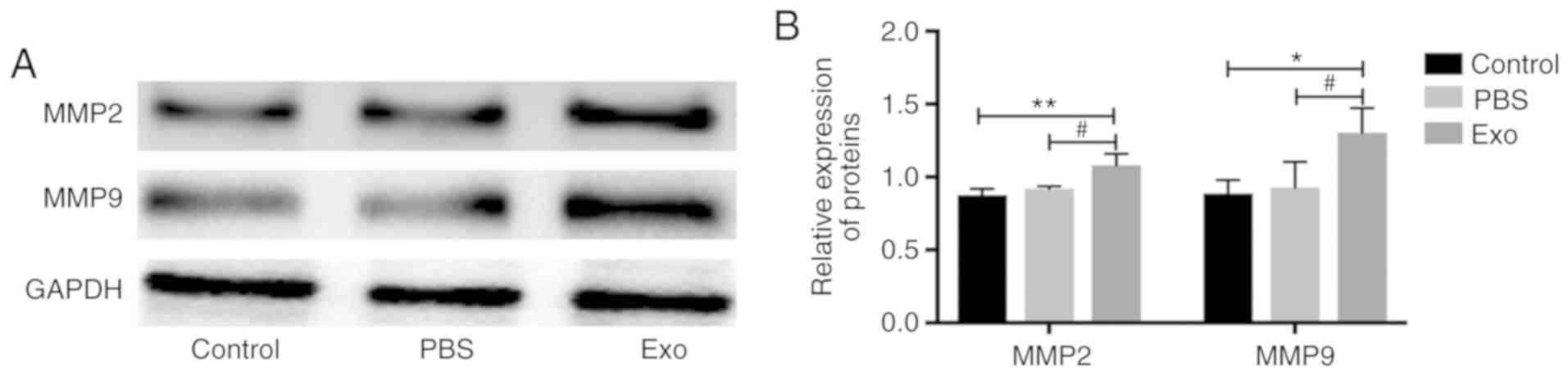
Piwil2-iCSC-Exo enhanced the
expression of MMP2 and MMP9 proteins in FBs
MMP2 and MMP9 are matrix mellatoproteases involved
in cancer progression and invasion and their expression is
considered a marker of cell invasiveness (38–41).
In order to investigate the likelihood that Piwil2-iCSC-Exo can
increase the invasive properties of FB cells, MMP2 and MMP9
expression levels were measured by western blot analysis.
Incubation with Piwil2-iCSC-Exo significantly increased the
expression of MMP2 (P<0.01 vs. the control group, P<0.05 vs.
the PBS group) and MMP9 (P<0.05) in comparison to the control
groups. No statistically significant differences were noted between
the control group and the PBS group (P>0.05; Fig. 4).
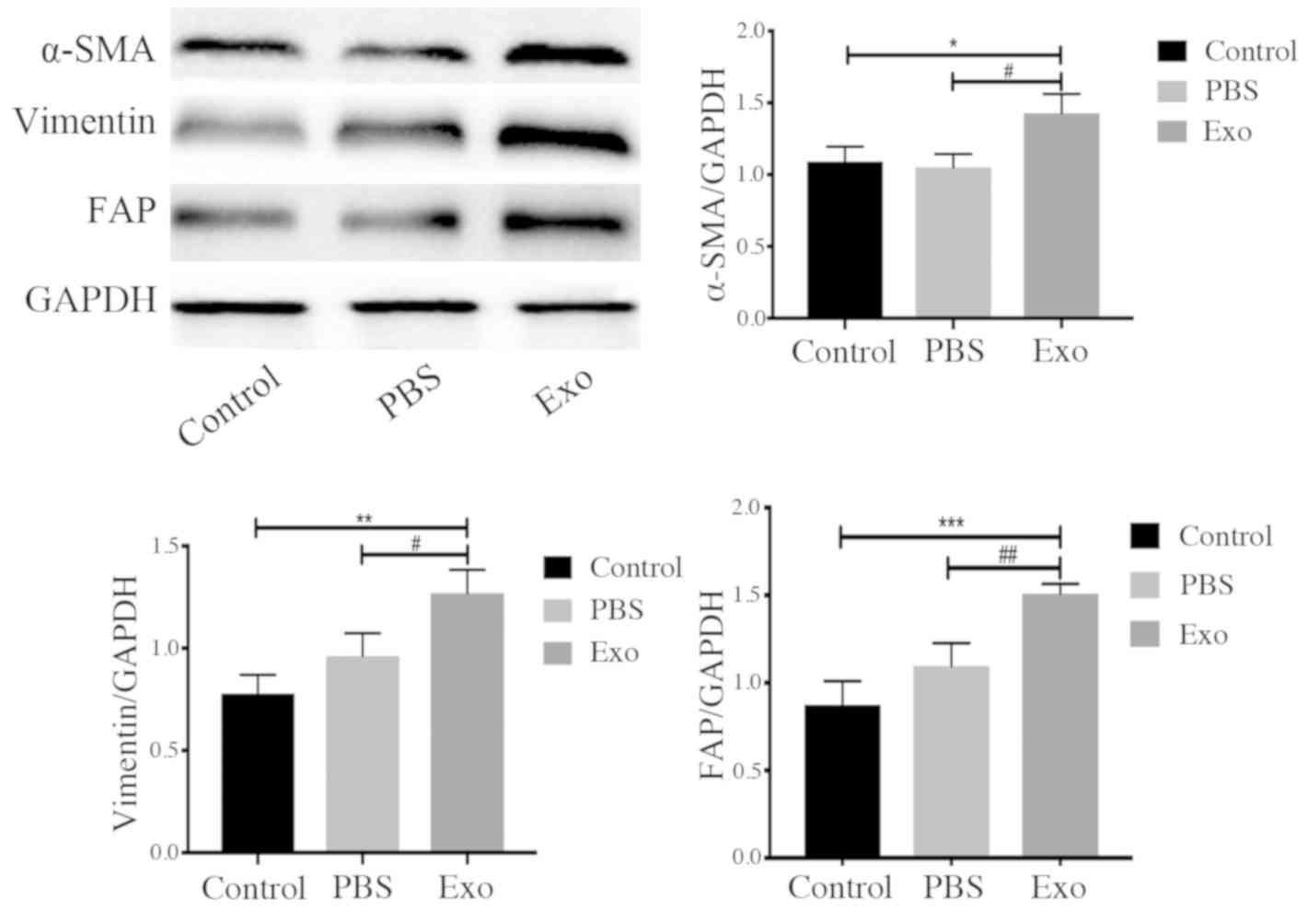
Piwil2-iCSC-Exo promote the
transformation of FBs into CAFs
CAFs are important components of the tumor
microenvironment and are involved in tumor development (42). CAFs are permanently activated
through the elevated expression of specific markers, such as FAP,
α-SMA and vimentin (43). As shown
in Fig. 5, incubation of the FBs
with Piwil2-iCSC-Exo increased the expression of α-SMA (P<0.05),
vimentin (P<0.01 vs. the control group, P<0.05 vs. the PBS
group) and FAP (P<0.001 vs. the control group, P<0.01 vs. the
PBS group) in comparison to the control groups.
Discussion
Previous studies have confirmed that CSCs are the
primary determinant of tumor initiation, propagation, metastasis
and recurrence (12–15). However, the exact mechanisms through
which CSCs promote tumor development remain unknown. A growing body
of data suggest that there is a strong association between exosomes
and tumor development (24–26). Exosomes, released by various types
of cells, can mediate the biological activities of recipient cells
through their contents, including proteins, nucleic acids and
lipids. Thus, it was hypothesized that CSC-derived exosomes may
promote tumor development by inducing changes in the tumor
microenvironment. The results of this study support this
hypothesis, as the CSC-derived exosomes significantly enhanced
proliferation, migration and invasion of FBs, a major cell-type of
the tumor microenvironment. These data suggest that the enhanced
migration and invasion may be caused by an increased
metalloproteinase activity in FBs. More importantly, it was
demonstrated that CSC-derived exosomes can induce the CAF-type
phenotype in stromal FBs.
Due in part to the low number of CSCs in tumors and
the difficulties encountered in the purification of CSCs from tumor
tissues, direct studies on native CSCs remain challenging.
Therefore, this study used Piwil2-iCSC reprogrammed cells,
transfected with the self-renewing gene Piwil2 (36), that served as a CSC model for in
vitro experiments. Using this model, it was demonstrated that
Piwil2-iCSC-Exo enhance the proliferation, migration and invasion
of FBs in vitro. Similar to the observations in this study,
previous studies have suggested that exosomes derived from advanced
tumors can enhance cell proliferation and cell motility (44–46).
Further evidence has confirmed that stem cell-derived exosomes can
carry pluripotent transcription factors, such as Nanog, Oct-4 and
Wnt family proteins, which promote the self-renewal of the
recipient cells by altering cell plasticity (47,48).
Sánchez et al demonstrated that human prostate CSC-derived
exosomes can promote the proliferation and migration of prostate
FBs through the activation of miR-139 (49). Another study revealed that thyroid
tumor stem cell-like exosomes promote distant metastasis formation
through non-coding RNAs (lncRNAs) (13). Furthermore, CD105+ renal
CSC-derived exosomes activate angiogenesis and promote lung
metastasis in vivo through the release of VEGF and MMP2
(50). Some studies have observed
that the tetraspanin family of proteins, such as CD9 and CD63, are
likely involved in cell proliferation, cell motility and metastasis
(51,52). It was thus speculated that the
bioactive substances contained in Piwil2-iCSC-Exo may alter the
biological behavior of FBs.
Metastasis formation depends on the ability of the
tumor cells to degrade the extracellular matrix and destroy the
basement membrane (2). MMPs are the
most effective proteases for degrading the extracellular matrix and
are implicated in multiple steps of tumorigenesis, as well as in
tumor invasion and metastasis formation (38,39).
MMP2 and MMP9 are the most distinctive MMPs characterized by a
strong proteolytic activity in the extracellular matrix (40,41).
Multiple studies have reported that MMP9 is overexpressed in tumor
cells and is thus linked to metastasis formation and ultimately, a
poor prognosis (53,54). Zhang et al (55) reported that the overexpression of
MMP9 can promote metastasis formation in non-small cell lung
cancer. It has also been shown that MMP2 and MMP9 may contribute to
metastasis in gastric adenocarcinoma (40). This study demonstrated that the
expression of MMP2 and MMP9 in FBs increased in the presence of
Piwil2-iCSC-Exo, an increase likely related to the invasiveness and
motility of the exposed cells.
CAFs are important components of the tumor
microenvironment and have been closely associated with the
occurrence and development of tumors. CAFs are constitutively
activated via the elevated expression of specific markers such as
α-SMA, vimentin, as well as FAP (42,43).
It has been shown that normal FBs can be induced to differentiate
into CAFs by exposure to their surrounding tumor cells (27,56).
In this study, the expression of α-SMA, vimentin and FAP was
increased in FBs exposed to Piwil2-iCSC-Exo, suggesting that
Piwil2-iCSC-Exo can induce the transformation of normal FBs into
CAFs. Taken together, it can be hypothesized that CSC-derived
exosomes can promote the development of tumors by altering the
composition of the tumor microenvironment.
In conclusion, the findings of this study provide
new evidence that exosomes derived from CSCs may promote tumor
progression and the development of distant metastasis by
transforming the composition of the tumor microenvironment. The
evidence presented herein confirms that exosomes play a role in
promoting FB proliferation, migration and invasion. The data
contribute to a better understanding of the complex association
between the CSCs and tumor development, which in turn could help to
develop a potential cancer-microenvironment targeted therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Special
Project of Science and Technology Innovation for Social
Undertakings and Livelihood Guarantee of Chongqing (grant no.
cstc2017shmsA130055 and no. cstc2017shmsA130103).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DZ and DL performed the experiments. DHu and BT
analyzed the results. WG, ZW, ZZ and LS performed the statistical
analysis. GW, DHe and DZ designed all the experiments. DZ wrote the
manuscript. LS, GW and DoH edited the manuscript. All authors
contributed to the revision of the manuscript and approved the
final version for publication. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CSCs
|
cancer stem cells
|
|
Piwil2-iCSCs
|
Piwil2 induced cancer stem cells
|
|
FBs
|
fibroblasts
|
|
Piwil2-iCSC-Exo
|
exosomes derived from Piwil2-iCSCs
|
|
CAFs
|
cancer-associated fibroblasts
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
PBS
|
phosphate-buffered saline
|
|
PVDF
|
polyvinylidene fluoride
|
|
TEM
|
transmission electron microscopy
|
|
NTA
|
nanoparticle tracking analysis
|
|
MMPs
|
matrix metalloproteinases
|
References
|
1
|
López-Gómez M, Malmierca E, de Górgolas M
and Casado E: Cancer in developing countries: The next most
preventable pandemic. The global problem of cancer. Crit Rev Oncol
Hematol. 88:117–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liao WT, Ye YP, Deng YJ, Bian XW and Ding
YQ: Metastatic cancer stem cells: From the concept to therapeutics.
Am J Stem Cells. 3:46–62. 2014.PubMed/NCBI
|
|
3
|
Kreso A and Dick J: Evolution of the
cancer stem cell model. Cell Stem Cell. 14:275–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ponti D, Costa A, Zaffaroni N, Pratesi G,
Petrangolini G, Coradini D, Pilotti S, Pierotti MA and Daidone MG:
Isolation and in vitro propagation of tumorigenic breast cancer
cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Costa FF, Le Blanc K and Brodin B: Concise
review: Cancer/testis antigens, stem cells, and cancer. Stem Cells.
25:707–711. 2010. View Article : Google Scholar
|
|
6
|
Kim KM, Calabrese P, Tavaré S and Shibata
D: Enhanced stem cell survival in familial adenomatous polyposis.
Am J Pathol. 164:1369–1377. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kucia M and Ratajczak MZ: Stem cells as a
two edged sword-from regeneration to tumor formation. J Physiol
Pharmacol. 57 (Suppl 7):S5–S16. 2006.
|
|
8
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Visvader JE: Cells of origin in cancer.
Nature. 469:314–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prasetyanti PR and Medema JP: Intra-tumor
heterogeneity from a cancer stem cell perspective. Mol Cancer.
16:412017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aguilar-Gallardo C and Simón C: Cells,
stem cells, and cancer stem cells. Semin Reprod Med. 31:005–013.
2013. View Article : Google Scholar
|
|
12
|
Phi LTH, Sari IN, Yang YG, Lee SH, Jun NY,
Kim KS, Lee KY and Kwon HY: Cancer stem cells (CSCs) in drug
resistance and their therapeutic implications in cancer treatment.
Stem Cells Int. 2018:54169232018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hardin H, Helein H, Meyer K, Robertson S,
Zhang RR, Zhong WX and Lloyd RV: Thyroid cancer stem-like cell
exosomes: Regulation of EMT via transfer of lncRNAs. Lab Invest.
98:1133–1142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valery AC, Golam K, Xia L, Mary D, Junk
DJ, Dongyin G, Chris H, Monica V, Erin MH and Maksim S: Cancer stem
cells: Targeting the roots of cancer, seeds of metastasis, and
sources of therapy resistance. Cancer Res. 75:924–929. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Albini A, Bruno A, Gallo C, Pajardi G,
Noonan DM and Dallaglio K: Cancer stem cells and the tumor
microenvironment: Interplay in tumor heterogeneity. Connect Tissue
Res. 56:414–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin WW and Karin M: A cytokine-mediated
link between innate immunity, inflammation, and cancer. J Clin
Invest. 117:1175–1183. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sittichai K: The tumor microenvironment
contribution to development, growth, invasion and metastasis of
head and neck squamous cell carcinomas. J Cancer. 4:66–83. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sund M and Kalluri R: Tumor stroma derived
biomarkers in cancer. Cancer Metastasis Rev. 28:177–183. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Paul G and Alexander S, Svenja R, Daniela
G, Steffen R, Condon TP, Alexander M, Minh-Chau P, Otwin L and
Alexander S: Cleavage of L1 in exosomes and apoptotic membrane
vesicles released from ovarian carcinoma cells. Clin Cancer Res.
11:2492–2501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hannafon BN and Ding WQ: Intercellular
communication by exosome-derived microRNAs in cancer. Int J Mol
Sci. 14:14240–14269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang S, Hu C, Liu P and Lu M:
Tumor-derived exosomes in cancer metastasis risk diagnosis and
metastasis therapy. Clin Transl Oncol. 21:152–159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abak A, Abhari A and Rahimzadeh S:
Exosomes in cancer: Small vesicular transporters for cancer
progression and metastasis, biomarkers in cancer therapeutics.
PeerJ. 6:e47632018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma A: Role of stem cell derived
exosomes in tumor biology. Int J Cancer. 142:2018. View Article : Google Scholar
|
|
27
|
Bhowmick NA, Neilson EG and Moses HL:
Stromal fibroblasts in cancer initiation and progression. Nature.
432:332–337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu Y, Yang W, Qin C, Zhang LF, Deng JJ,
Liu SB and Qin ZH: Responsiveness of stromal fibroblasts to
IFN-gamma blocks tumor growth via angiostasis. J Immunol.
183:6413–6421. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao JX: Cancer stem cells: The lessons
from pre-cancerous stem cells. J Cell Mol Med. 12:67–96. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li D, Sun X, Yan D, Huang J, Luo Q, Tang H
and Peng Z: Piwil2 modulates the proliferation and metastasis of
colon cancer via regulation of matrix metallopeptidase 9
transcriptional activity. Exp Biol Med (Maywood). 237:1231–1240.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K,
Jarjoura D, Nakano T, Ramesh GK, Shapiro CL, et al: Piwil2 is
expressed in various stages of breast cancers and has the potential
to be used as a novel biomarker. Int J Clin Exp Pathpl. 3:328–337.
2010.
|
|
32
|
He G, Chen L, Ye Y, Xiao Y, Hua K,
Jarjoura D, Nakano T, Barsky SH, Shen R and Gao JX: Piwil2
expressed in various stages of cervical neoplasia is a potential
complementary marker for p16. Am J Transl Res. 2:156–169.
2010.PubMed/NCBI
|
|
33
|
Chen C, Liu J and Xu G: Overexpression of
PIWI proteins in human stage III epithelial ovarian cancer with
lymph node metastasis. Cancer Biomark. 13:315–321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Liu Y, Shen X, Zhang X, Chen X,
Yang C and Gao H: The PIWI protein acts as a predictive marker for
human gastric cancer. Int J Clin Exp Pathol. 5:315–325.
2012.PubMed/NCBI
|
|
35
|
Greither T, Koser F, Kappler M, Bache M,
Lautenschläger C, Göbel S, Holzhausen HJ, Wach S, Wurl P and
Taubert H: Expression of human Piwi-like genes is associated with
prognosis for soft tissue sarcoma patients. BMC Cancer. 12:2722012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang D, Wu X, Liu X, Cai C, Zeng G,
Rohozinski J, Zhang Y, Wei G and He D: Piwil2-transfected human
fibroblasts are cancer stem cell-like and genetically unstable.
Oncotarget. 8:12259–12271. 2017.PubMed/NCBI
|
|
37
|
Xue M, Chen W, Xiang A, Wang R, Chen H,
Pan J, Pang H, An H, Wang X, Hou H and Li X: Hypoxic exosomes
facilitate bladder tumor growth and development through
transferring long non-coding RNA-UCA1. Mol Cancer. 16:1432017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deryugina EI and Quigley JP: Tumor
angiogenesis: MMP- mediated induction of intravasation- and
metastasis-sustaining neovasculature. Matrix Biol. 44-46:94–112.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yamamoto K, Murphy G and Troeberg L:
Extracellular regulation of metalloproteinases. Matrix Biol.
44-46:255–263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang Q, Lan F, Wang X, Yu Y, Ouyang X,
Zheng F, Han J, Lin Y, Xie Y, Xie F, et al: IL-1β-induced
activation of p38 promotes metastasis in gastric adenocarcinoma via
upregulation of AP-1/c-fos, MMP2 and MMP9. Molecular cancer.
13:182014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu J, Ping W, Zu Y and Sun W:
Correlations of lysyl oxidase with MMP2/MMP9 expression and its
prognostic value in non-small cell lung cancer. Int J Clin Exp
Pathol. 7:6040–6047. 2014.PubMed/NCBI
|
|
42
|
Shimoda M, Mellody KT and Orimo A:
Carcinoma-associated fibroblasts are a rate-limiting determinant
for tumour progression. Semin Cell Dev Biol. 21:19–25. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu R, Li H, Liu L, Yu J and Ren X:
Fibroblast activation protein: A potential therapeutic target in
cancer. Cancer Biol Ther. 13:123–129. 2015. View Article : Google Scholar
|
|
44
|
Wu L, Zhang X, Zhang B, Shi H, Yuan X, Sun
Y, Pan Z, Qian H and Xu W: Exosomes derived from gastric cancer
cells activate NF-κB pathway in macrophages to promote cancer
progression. Tumour Biol. 37:12169–12180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gangoda L, Liem M, Ang CS, Keerthikumar S,
Adda CG, Parker BS and Mathivanan S: Proteomic profiling of
exosomes secreted by breast cancer cells with varying metastatic
potential. Proteomics. 17:2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sakha S, Muramatsu T, Ueda K and Inazawa
J: Exosomal microRNA miR-1246 induces cell motility and invasion
through the regulation of DENND2D in oral squamous cell carcinoma.
Sci Rep. 6:387502016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tooi M, Komaki M, Morioka C, Honda I,
Iwasaki K, Yokoyama N, Ayame H, Izumi Y and Morita I: Placenta
mesenchymal stem cell derived exosomes confer plasticity on
fibroblasts. J Cell Biochem. 117:1658–1670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Julia Christina G, Varun C, Kerstin B and
Michael B: Active Wnt proteins are secreted on exosomes. Nat Cell
Biol. 14:1036–1045. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sánchez CA, Andahur EI, Valenzuela R,
Castellón EA, Fullá JA, Ramos CG and Triviño JC: Exosomes from bulk
and stem cells from human prostate cancer have a differential
microRNA content that contributes cooperatively over local and
pre-metastatic niche. Oncotarget. 7:3993–4008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Grange C, Tapparo M, Collino F, Vitillo L,
Damasco C, Deregibus MC, Tetta C, Bussolati B and Camussi G:
Microvesicles released from human renal cancer stem cells stimulate
angiogenesis and formation of lung premetastatic niche. Cancer
Research. 71:5346–5356. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Berditchevski F: Complexes of tetraspanins
with integrins: More than meets the eye. J Cell Sci. 114:4143–4151.
2001.PubMed/NCBI
|
|
52
|
Mantegazza AR, Barrio MM, Moutel S, Bover
L, Weck M, Brossart P, Teillaud JL and Mordoh J: CD63 tetraspanin
slows down cell migration and translocates to the
endosomal-lysosomal-MIICs route after extracellular stimuli in
human immature dendritic cells. Blood. 104:1183–1190. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nart D, Yaman B, Yilmaz F, Zeytunlu M,
Karasu Z and Kiliç M: Expression of matrix metalloproteinase-9 in
predicting prognosis of hepatocellular carcinoma after liver
transplantation. Liver Transpl. 16:621–630. 2010.PubMed/NCBI
|
|
55
|
Zhang W, Zhang T, Lou Y, Yan B, Cui S,
Jiang L and Han B: Placental growth factor promotes metastases of
non-small cell lung cancer through MMP9. Cell Physiol Biochem.
37:1210–1218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yu L, Yang W, Qin C, Zhang L, Deng J, Liu
S and Qin Z: Responsiveness of stromal fibroblasts to IFN-gamma
blocks tumor growth via angiostasis. J Immunol. 183:6413–6421.
2009. View Article : Google Scholar : PubMed/NCBI
|