Under physiological and pathological conditions,
cell life cycle comes to an end with cell death. Necrosis and
apoptosis are considered forms of cell death. Studies have reported
that certain forms of programmed cell death, such as autophagy and
necroptosis, exhibit unique pathophysiological features that are
distinct from other forms of programmed cell death, such as
necrosis and apoptosis (1,2). In 2012, the concept of ferroptosis
was proposed (3). During
ferroptosis, lipid reactive oxygen species (ROS) are produced as a
result of iron-dependent, non-apoptotic cell death. In ferroptosis,
iron reacts with hydrogen peroxide, releasing electrons and
generating hydroxyl radicals. These reactions damage intracellular
lipids and proteins and cause oxidative damage to DNA, which
accelerates death of tumor cells (4). Ferroptosis is morphologically and
functionally distinct from necrosis, autophagy and apoptosis; it
does not show typical characteristics of necrosis or apoptosis,
including cytoplasmic swelling, cell shrinkage and rupture,
presence of apoptotic bodies or cytoskeletal disintegration.
Additionally, unlike autophagy, it does not involve formation of
closed lipid membrane bilayers, but a specific structure known as
the autophagic vacuole (5). In
terms of morphology, ferroptosis is marked by mitochondrial
shrinkage and increased membrane density (6). Ferroptosis is associated with a range
of diseases (eg:gastric cancer) (7). Ferroptosis is regulated by epigenetic
mechanisms (8). Epigenetic
modifications are alterations in gene expression or cell phenotype
caused by heritable changes that affect DNA methylation, histone
modifications and non-coding RNA (ncRNA) regulation. Epigenetic
mechanisms also control gene transcription, cell proliferation,
developmental processes and immune function (9). Furthermore, epigenetic regulation is
implicated in ferroptosis and tumorigenesis via modulation of
metabolic genes and intermediates, thereby regulating and altering
lipid peroxidation (10). The
epigenetic control of ferroptosis also provides novel directions
for development of therapeutic interventions, which may overcome
the current barriers in antitumor therapy (11). The present review aimed to discuss
how epigenetic regulators regulate ferroptosis and how ferroptosis
contributes to tumor biology.
Iron is a key trace element in the human body. It
participates in heme synthesis and serves an important role in
various physiological metabolic processes as a co-factor of certain
key enzymes (12). The human body
maintains iron homeostasis by regulating intestinal circulation and
absorption of iron in the reticuloendothelial system via
coordination of iron absorption, utilization, storage and
circulation (13). There are
numerous genes involved in maintaining iron homeostasis. Cellular
iron homeostasis is primarily controlled by iron regulatory protein
(IRP)-iron response element (14).
IRP is involved in regulation of iron absorption, transport and
storage, alongside transferrin receptor 1 (TFR1), divalent metal
ion transporter (DMT1) and ferritin (15). Iron metabolism in the human body is
primarily regulated by hepcidin-ferroportin (HAMP-FPN), in which
HAMP regulates iron uptake and release in tissue, as well as iron
homeostasis in the body (16).
Under iron overload, HAMP binds to FPN on target cell membranes and
internalizes and degrades FPN in lysosomes, thereby inhibiting iron
absorption by enterocytes and releasing iron from macrophages or
hepatocytes into the serum, thus regulating iron absorption and
distribution (17). At the
molecular level, HAMP is regulated by the bone morphogenetic
protein (BMP)/hemojuvelin (HJV)/SMAD signaling pathway (18). HJV is a co-receptor of BMP
(19). By enhancing BMP
expression, the SMAD signaling pathway is stimulated and HAMP
expression is promoted (20). BMP
stimulates HAMP expression; this process is also regulated by HFE
protein (21). In a high-iron
environment, transferrin competes to capture HFE-bound TFR1,
resulting in HFE dissociating from TFR1 and binding to TFR2,
thereby activating the SMAD signaling pathway and inducing HAMP
expression. Membrane-type serine protease 2 inhibits HAMP
expression by shear HJV negative feedback. Any gene abnormalities
in the signal transduction pathway regulated by HAMP affect the
expression of HAMP and cause disorders associated with iron
metabolism (22). Recent studies
(23–25) on iron metabolism have revealed that
epigenetic inheritance serves an important role in iron homeostasis
metabolism. DNA methylation generally occurs in cytosine-guanine
dinucleotide CpG-rich DNA regions, called CpG islands (CGIs), which
are the most widely studied epigenetic modifications and serve an
important role in the regulation of gene expression. Vertebrates
have a higher methylation rate of CpG dinucleotides than mammals,
with ~80% containing 5-methylcytosine (5mC), a gene silencing
marker. There are certain exceptions to this hypermethylated state,
namely CGIs, which are typically hypomethylated in comparison with
the rest of the genome (26). CGIs
in promoter regions overlap in the earliest vertebrates and humans,
suggesting a coevolutionary CGI-compatible system. DNA sequence
features are characteristic of CGI, including hypomethylation of
DNA, high CpG and GC content and transcription factor binding
(27). Transcription factors and
chromatin-modifying enzymes are recruited along with
transcriptional activators by these sequence features. CGIs
represent a ubiquitous class of DNA sequences commonly associated
with promoters of vertebrate genes whose sequence features make
them transcriptionally active (28). DNA sequence and chromatin
determinants allow identification of CGIs, including decreased DNA
methylation (5mC), increased CpG and GC content and histone H3
trimethylation (H3K4me3) and transcription factor binding sites
(29). Increased DNA methylation
leads to gene silencing, while decreased DNA methylation activates
gene expression. Iron metabolism is associated with DNA methylation
and iron level and Oxidation state affect DNA methylation (30). Increased free iron content in the
brain due to oxidative stress leads to excessive S-adenosine
homocysteine in HFE mutant mice by inhibition of the activity of
DNA methyltransferase (31).
Treatment with the iron chelator deferoxamine improves activity of
DNA methyltransferase and certain breast cancer cells are induced
by methylation (32).
3-Hydroxybutyrate dehydrogenase 2 is a rate-limiting enzyme in
mammalian ferriphilin synthesis. Inhibition of its expression
causes iron overload in cells and mitochondria (33). In tumor cell proliferation, iron
demand is increased and the expression of genes associated with
iron metabolism is affected. Previous studies (34,35)
have found that low expression of FPN in patients with breast
cancer indicates poor survival. In vitro studies (36,37)
also found that inhibition of FPN expression promotes tumor growth,
while overexpression of FPN hinders tumor growth. Low expression of
FPN in breast cancer is regulated by nuclear factor erythroid
2-related factor 2 (NRF2) and is inhibited by hypermethylation of
CGIs in the FPN promoter region, indicating that breast cancer can
be inhibited by targeting FPN or upstream regulatory factors
(38). HAMP expression is
inhibited in patients with hepatocellular carcinoma (HCC), which is
accompanied by hypermethylation of the highly conserved CGI site in
its promoter region (38).
Demethylating drug treatment removes hypermethylation of the HAMP
promoter and upregulates expression of HAMP in HCC cells (39). TFR2 in HCC cells is also regulated
by hepcidin in hepatocellular carcinoma. Therefore, regulation of
HAMP by hypermethylation is a recently (40) identified form of epigenetic
regulation of iron metabolism, which provides a novel area of
research on tumors and iron. Since iron can easily change its
valence state and switch between Fe2+ and
Fe3+, iron is a key catalyst to produce active free
radicals within aerobic organisms. Rigid binding between iron and
transferrin occurs in vivo. The activity of O2−
and H2O2 is not sufficient for oxidation of
certain macromolecules, such as nucleic acids and proteins. Iron
overload is not always toxic and the excess iron can initially be
chelated in ferritin or lysosomes in a safe manner. However, once
the quantity of accumulated iron exceeds storage capacity, Iron not
chelated with ferritin emerges, promoting oxidative stress
responses (41). The reaction
between Fe2+ and H2O2 produces
hydroxyl radical (HO.), which is known as the Fenton reaction
(42). Iron not chelated with
ferritin is more conducive to Fenton reaction than iron chelated
with ferritin and unstable iron present in the body passes through
HO. with RO. generation, which enhances the effect of
H2O2 and other organic peroxides in cells
(43). The majority of
O2 consumed by aerobic organisms is safely reduced to
H2O in a four-electron transfer reaction catalyzed by
the cytochrome oxidase complex IV of the mitochondrial inner
membrane respiratory chain (44).
However, a fraction of unused O2 is involved in other
physiological reactions, such as phagocytosis and immune
activation. The toxic intermediates that form are called ROS. Under
normal conditions, specific enzymes rapidly metabolize ROS, such as
glutathione peroxidase (GPX) (45). There are a variety of biochemical
and physiological oxidative processes in the body that produce ROS;
these processes are also linked to numerous physiological and
pathological processes, eg:heart disease (46). By regulating intracellular signal
transduction and homeostasis, ROS exert beneficial effects at low
concentrations. At high concentrations, however, ROS primarily
cause protein, lipid and DNA damage. Endogenous ROS are primarily
derived from byproducts of subcellular organelles such as
mitochondria. A normal cell can become malignant if it contains
high levels of ROS. It has been shown that ROS influence epigenetic
inheritance, including DNA methylation (47). In cancer cells, ROS enhance DNA
methylation, leading to silencing of tumor suppressor and
antioxidant genes, and cancer cell proliferation under oxidative
stress. In colorectal cancer cells, ROS increase expression of
caudal type homeobox 1 (CDX1) (48). Treatment with
H2O2 increases expression and activity of
CDX1, DNA methylated transferase 1 (DNMT1) and histone deacetylase
(HDAC1) in cancer cells, which promotes cancer progression
(49). In Parkinson's disease,
peroxisome proliferators-activated receptors coactivator 1α
(PGC-1α) downregulation is associated with mitochondrial
dysfunction, oxidative stress and inflammation. PGC-1α is required
by nerve cells to induce proteins that detoxify ROS, such as GPX
(50). In PGC-1α knockout mice,
glial cells are more susceptible compared with normol mice to
neuroinflammation induced by Toll-like receptor 4 agonists. PGC-1α
downregulation leads to increased ROS and neurological damage
(51,52) Previous studies (53,54)
have shown that proinflammatory fatty acid palmitate leads to
atypical cytosine methylation in the PGC-1α gene promoter and
decreases gene expression and mitochondrial content. Thus, PGC-1α
promoter methylation is associated with dysfunctional inflammatory
signaling (55). Cells in the body
continuously produce and remove ROS in a dynamic equilibrium under
physiological conditions. Once redox imbalance occurs in cells,
redox signals are generated, which lead to adaptive changes in gene
expression. Iron and iron derivatives bind to ROS-producing
enzymes, such as the NADPH and cytochrome P450 enzyme systems.
Lysosomes also contain redox-active iron pools from extracellular
sources that catalyze the generation of damaging free radicals via
the Fenton reaction (56).
Cell death mediated by ferroptosis occurs in
numerous types of disease, including cancer. Unlike autophagy and
apoptosis, ferroptosis is characterized by dependence on
intracellular free iron ions and ROS (57). Activated circulating iron
(Fe3+) enters cells and is reduced to Fe2+ by
ferrooxidoreductase six transmembrane epithelial antigen 3
(58). Once endosomes release
Fe2+ via DMT1, it is transported to the cytoplasm, where
it couples with ROS, leading to lipid peroxidation and ferroptosis
(59). ROS are molecules
containing reduced oxygen, which include superoxide
(O2−) and peroxides (H2O2 and
ROOH), as well as free radicals (HO and RO). In cells, increased
ROS production induces ferroptosis (60). In cases of lipid peroxidation and
oxidative stress, the permeability of the cell membrane directly
leads to reduction or disappearance of intracellular mitochondria
and the rupture or aggregation of mitochondrial membranes (61). In the case of malignant tumors,
ferroptosis serves a crucial role in eradicating tumor cells as an
adaptive process (62). Tumor
cells are highly sensitive to ferroptosis, which controls the
abundance of iron by activating expression of transferrin receptor
and ferritin, thus activating the RAS/MEK signaling pathway. When
the RAS/MEK signaling pathway is activated, tumor cells are
prevented from absorbing cystine and ROS are released through
mitochondrial voltage-dependent anion channels (63). For example, activated RAS/MEK
signaling induces expression of SOSC1, which controls expression of
P53, decreases expression of cystine transporter and sensitizes
cells to ferroptosis (64). In
addition, various components (Beclin1) promote ferroptosis and
indirectly decrease activity of system Xc (65). Low member 11 of the solute carrier
family 7 (SLC7A11) expression prevents Cys2 transport and
activation of the glutathione (GSH)-independent thioredoxin (TXN)
system, leading to dysfunction of the GSH/GPX4 lipid peroxidation
pathway (66). Mechanistically,
this may be due to the inability to transfer a sufficient quantity
of reducing equivalents from Cys2 to TXN (via TXN reductase) to
maintain the endogenous antioxidant lipid α-tocopherol in a reduced
state, thereby promoting ROS accumulation and ferroptosis (67). In cancer, the mechanism of
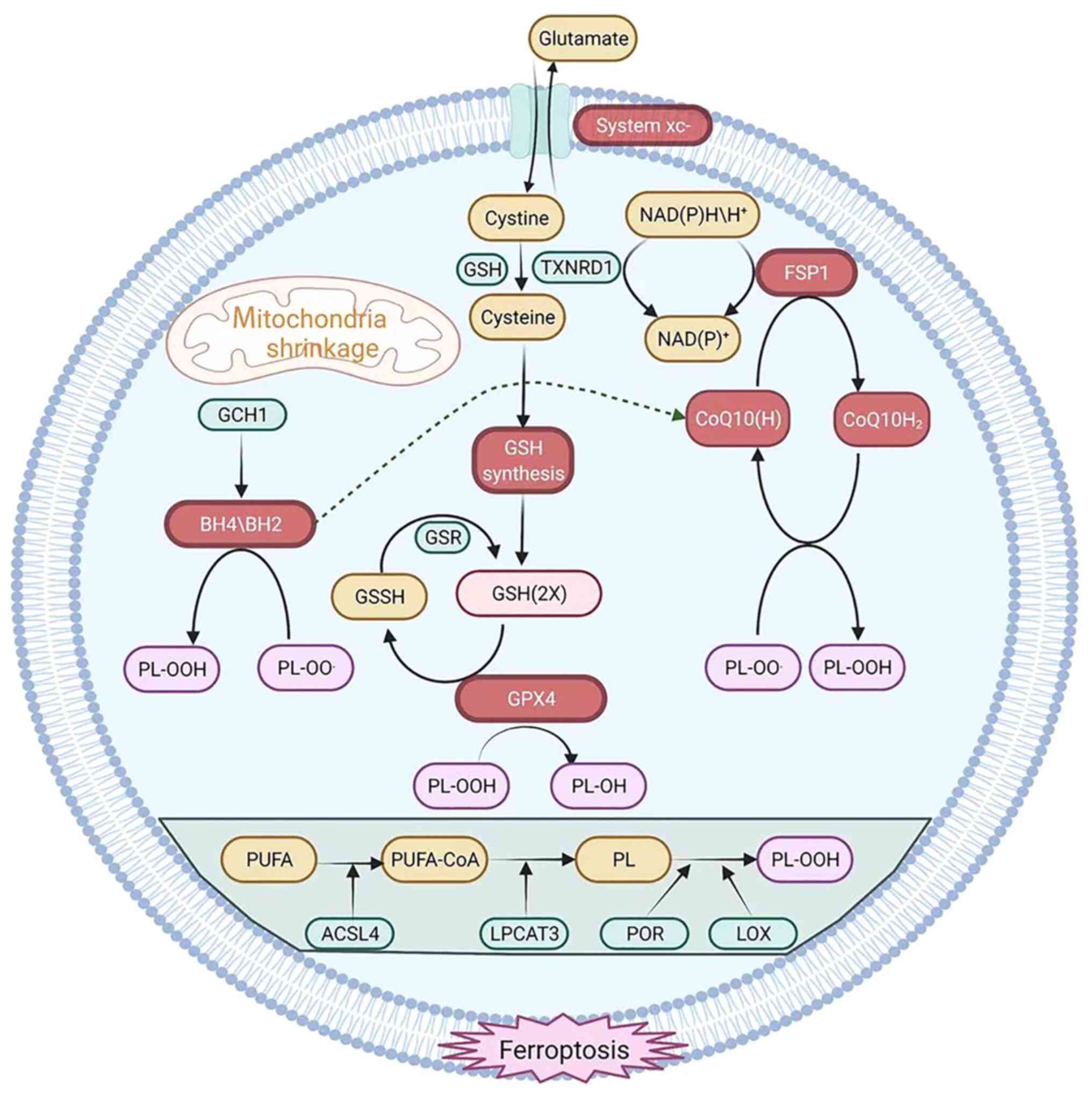
ferroptosis primarily involves three major systems (Fig. 1).
The antioxidant system Xc- consists of transporters
located on cell membranes. The primary system maintains homeostasis
of glutamate and cystine on cell membranes, where it pumps out
glutamate molecules and pumps in cystine molecules (68). The functional subunits of the
system Xc- are SLC7A11 and SLC3A2 (68). GSH is synthesized by reduction of
cystine to cysteine by system Xc-. Glutamyl-L-cysteine-L-glycine
(GSH) is an endogenous antioxidant consisting of glutamic acid,
cysteine and glycine that scavenges free radicals. Its production
is dependent on activity of glutamate-cysteine ligase and
inhibition of system Xc- attenuates its activity, resulting in GSH
depletion and leading to ferroptosis (69). Trans-sulfuration is another
mechanism by which methionine synthesizes cysteine. Cysteine is
still synthesized when intracellular system Xc- is inhibited
(70). Thus, ferroptotic inducers
that negatively regulate system Xc- cannot effectively kill cells.
The trans-sulfuration pathway is activated by interfering with RNA
dynamics to decrease expression of cysteine transfer RNA
synthetase. As a result, cells become less sensitive to
ferroptosis-inducing agents (71).
Cells that contain GPX4 decrease lipid peroxidation, which promotes
their survival. Aberrant expression of GPX4 is linked to a range of
diseases (breast cancer) in humans and depletion of GPX4 increases
H2O2-containing phospholipids (72), promotes lipoxygenase-mediated lipid
peroxidation and induces ferroptosis (73). GPX4 is highly expressed in tumor
tissue, while histone H3 is trimethylated at lysine 4 (H3K4me3) and
acetylated at lysine 27 (H3K27ac) at the GPX4 transcriptional start
site. Patients with cancer have a poor prognosis when epigenetic
modifications of GPX4 are present; thus, elevated expression of
GPX4 in tumor tissue may be due to epigenetic modifications
(74). In addition, the activation
of autophagy degrades intracellular ferritin and directly induces
ferroptosis in tumor cells. Autophagy is a lysosome-dependent
process that promotes ferroptosis by generating lysosomal ROS.
Promoting autophagy effectively enhances ferroptosis in tumor cells
(75). The autophagy-mediated
nuclear receptor coactivator (NCOA4) signaling pathway promotes
ferritin degradation and enhances ferroptosis when NCOA4 is
overexpressed (76), which
demonstrates the association between ferroptosis and apoptosis.
Although GPX4 is a key regulator of ferroptosis,
inhibition of GPX4 does not induce ferroptosis in certain cell
lines. A previous study identified potential factors that regulate
ferroptosis independently of the GPX4 signaling pathway (72). Cells were treated with a
ferroptosis initiator [RAS selective lethal 3 (RSL3)], and it was
observed that Glutathione Independent Iron Death Inhibitor Protein
(FSP1) induced apoptosis. The inhibition of GPX4 leads to
ferroptosis when FSP1 is overexpressed. Therefore, FSP1 may act as
a ferroptosis inhibitor (77).
However, AIFM1, although homologous to FSP1, does not inhibit
ferroptosis (78). FSP1 may
inhibit ferroptosis because its N-terminal myristoylated mediates
FSP1 localization to the plasma membrane. Furthermore,
myristoylation of the N-terminus of FSP1 promotes binding of target
proteins to the cell membrane. In the plasma membrane, COQ10 serves
as a lipophilic radical scavenger (79), while FSP1 serves as a
NADPH-dependent COQ oxidoreductase that regulates COQ10 in
vitro. FSP1 is a component of the traditional mitochondrial
respiratory chain that catalyzes the same reactions as complex I.
Extra-mitochondrial ubiquinone is reduced from COQ10 by FSP1, which
either directly captures lipid free radicals or indirectly serves
as an antioxidant by recycling α-tocopherol (77). Idebenone, a soluble analog of
COQ10, inhibits ferroptosis and lipid peroxidation by catalyzing
the first step in the biosynthesis of COQ2 (80). The aforementioned findings showed
that FSP1, like GPX4, inhibits ferroptosis by regulating the
non-mitochondrial COQ10 antioxidant system (80).
The redox-active cofactor BH4 serves a role in
production of neurotransmitters, carbon monoxide and aromatic amino
acids. In vitro, BH4 exhibits antioxidant properties and
GCH1 limits BH4 synthesis (82).
In tumor cells, GCH1 overexpression clears lipid peroxidation and
protects cells from ferroptosis. Overexpression of GCH1 protects
against RSL3-induced ferroptosis when cells were treated with RSL3
and apoptosis-inducing agents. GCH1 protects cells from ferroptosis
(83). Treatment of RSL3-exposed
cells with BH4 completely prevents ferroptosis. It has been
previously shown that GCH1 acts independently of the ferroptosis
and GSH pathways to prevent ferroptosis (84). Notably, BH4 as a cofactor for
biosynthetic enzymes does not serve a role in ferroptosis
protection (85). In addition, BH4
converts phenylalanine to tyrosine to promote synthesis of COQ10,
thereby exerting an antioxidant effect (86).
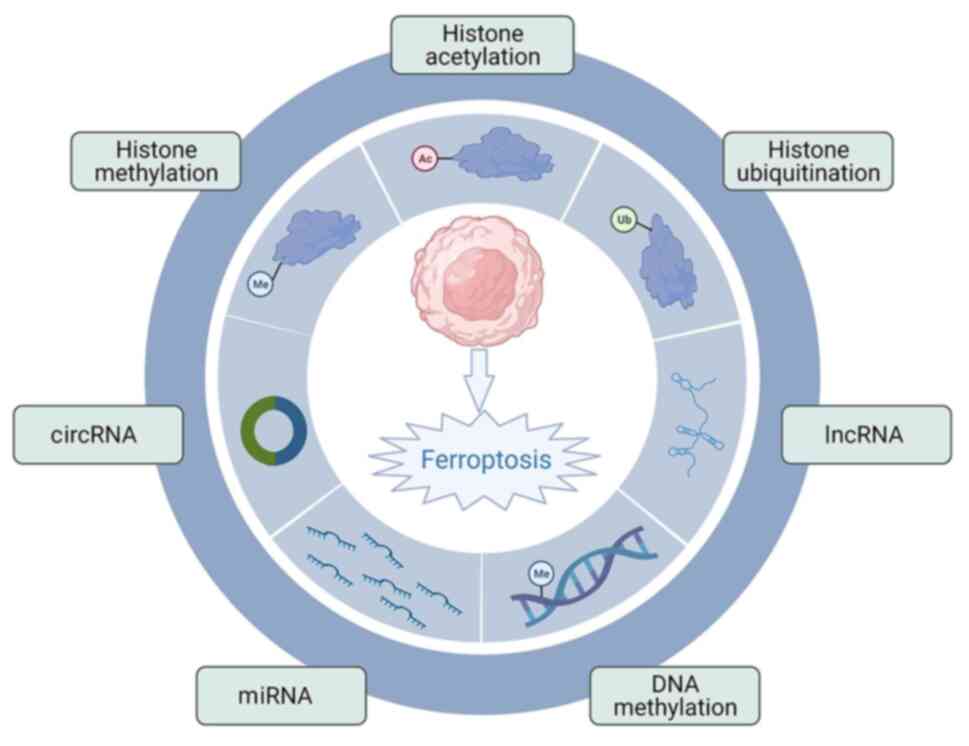
Alterations in DNA methylation, histone
modifications and ncRNA regulation are characteristic of epigenetic
regulation, which determines gene transcription, cell
proliferation, developmental processes and immune function
(87). Epigenetic regulation is
important for ferroptosis.
There are numerous roles for DNA methylation in
cancer, including suppression of DNA methylation at transcriptional
start sites of key gene regulatory elements, such as enhancers and
promoters (88). In DNA
methylation, a methyl group is typically inserted at the carbon 5
position of the cytosine base (5mC) via DNA methylation mediated by
CpG transferase DNMTs (89). DNA
methylation primarily regulates mitotic gene expression, centromere
stability and chromatin segregation. In addition,
5-hydroxymethylcytosine (5hmC) has been demonstrated to be produced
by 5mC oxidation. 5hmC has been widely studied in relation to its
potential role in modifying methylation landscapes by ten-eleven
translocation (TET) enzymes (90,91).
The ability of TET proteins to oxidize 5hmC to 5-formylcytosine and
5-carboxycytosine indicates utilization of the base excision repair
pathway, while thymine DNA glycosylase excises cytosine and
replaces it with an unmodified cytosine (92). CpG sites are found at ~28 million
sites in the human genome. The distribution of CpG sites in somatic
cells is uneven and ~70% of CpG sites are oxidized in normal
somatic cells (93). Clusters of
CpG sites are known as CGIs; CpG sites in CGIs tend not to be
methylated in somatic cells. Chromosomes are their primary medium.
Unmethylated CpG sites are found in promoter CGIs, where
transcription factors bind to control gene expression. The CpG
coast is ~2 kb pairs away from the CGI and has a lower density of
CpGs than the CGI (94). Normal
cells also contain these unmethylated regions that regulate gene
activity (95). During
tumorigenesis, normal epigenetic processes such as DNA methylation
are disrupted. Tumor development is primarily accompanied by
genome-wide hypomethylation and DNA hypermethylation in the
promoter regions of CGIs (96).
Cell proliferation and tumor suppressor genes and downstream
signaling pathways are primarily silenced by the hypermethylation
of CGIs (97). Moreover, tumors
also abnormally express DNMT enzymes, resulting in aberrant DNA
methylation across the genome, potentially causing mutations in
genomic sequences (98).
Aneuploidy and genomic instability are associated with genome-wide
hypomethylation in cancer (99).
The abnormal expression of transposable elements and oncogenes is
also associated with genome-wide DNA hypomethylation, which
disrupts cellular pathways and changes chromatin structure, as
chromatin structure and DNA methylation are associated and a
nucleosome is required before 5mC can be obtained (100). To anchor the DNMT enzyme,
abnormal DNA methylation must disrupt chromatin structure (101). The most studied (102) epigenetic modification is DNA
methylation (103). However, the
mechanism of this process in unclear and the number of
cancer-associated genes that are silenced and targeted is unknown.
A recent study on the association between DNA methylation,
ferroptosis and cancer has led to novel targeted therapy for cancer
(104). Hydricase, lymphoid
specific (HELLS/LSH) is a chromatin remodeling enzyme that inhibits
ferroptosis by activating metabolic genes such as stearoyl-coenzyme
A desaturase (SCD). HELLS induces epigenetic silencing of the long
non-coding RNA (lncRNA) LINC00472. LINC00472, as a tumor
suppressor, is downregulated in tumors and can inhibit ferroptosis
(105). Egl nine homolog 1 and
c-Myc in tumor cells activate LSH expression by inhibiting hypoxia
inducible factor 1. LSH inhibits iron dynamics by activating GLUT1,
SCD1 and fatty acid desaturase 2, which are involved in lipid
metabolism, by interacting with WD repeat domain 76. Several types
of cancer tissue (pancreatic and lung cancer) express high levels
of SCD1, an enzyme that catalyzes fatty acid synthesis at the
rate-limiting step. Through inhibition of SCD1, COQ10 activity is
decreased, resulting in lipid oxidation and cell death (106). GPX4 DNA methylation in the
nucleus pulposus is induced by homocysteine treatment, resulting in
ferroptosis (107). Furthermore,
DNA hypermethylation of the cadherin 1 (CDH1) gene promoter
increases ferroptosis by suppressing expression of E-cadherin,
which is encoded by CDH1 (108).
DNA methylation contributes to ferroptosis-associated gene
silencing, as demonstrated by the aforementioned studies. There is,
however, a need for further research to determine whether DNA
methylation also affects other ferroptosis-associated genes.
Notably, TET proteins also catalyze DNA demethylation by oxidizing
5mC (109).
RNA is a key biological macromolecule in cells and
it can be classified into two categories according to its function,
namely, coding RNA and ncRNA. RNA is responsible for gene
regulation and information transmission and is involved in disease.
For example, nc-RNAs encode tumor peptides or proteins (110,111). The following subsections review
ncRNAs in the context of microRNAs (miRNAs or miRs), lncRNAs and
circular RNAs (circRNAs) involved in ferroptosis.
RNA molecules >200 nucleotides in length are
known as lncRNAs and influence gene expression at different levels,
including transcription and post-transcriptional translation.
lncRNAs have also been reported to be key for a variety of
biological processes, such as cell differentiation, regulation of
the cell cycle and maintenance of stem cell pluripotency (127). Via downregulation of membrane
receptors and inhibition of necroptosis-associated proteins,
overexpression of lncRNAs inhibits the extrinsic apoptotic pathway
in tumor cells (128). lncRNAs
are important regulators of ferroptosis. A number of them affect
miRNAs, while others bind to specific enzymes. Wang et al
(129) reported that the lncRNAs
LINC00336 and embryonic lethal, abnormal vision, Drosophila-like 1
(ELAVL1) inhibit ferroptosis by interacting with each other. In
addition, ELAVL1 increases LINC00336 expression by stabilizing its
post-transcriptional levels when RSL3 is used to activate
ferroptosis in lung cancer cell lines. Overexpression of LINC00336
inhibits ferroptosis caused by RSL3 treatment, as well as protein
kinase-induced ferroptosis. Furthermore, overexpression of
LINC00336 decreases intracellular Fe2+ and mitochondrial
superoxide concentrations, as well as ROS production, demonstrating
that LINC00336 overexpression inhibits ferroptosis (129). Urothelial cancer associated 1
(UCA1) sponges miR-16 and increases expression of glutaminase 2
(GLS2) in bladder cancer cells. There are two mechanisms by which
lncRNAs affect ferroptosis (130). GLS2 enzyme catalyzes conversion
of glutamine to glutamate and subsequent synthesis of GSH. NADPH is
produced when glutamine enters the tricarboxylic acid cycle.
Glutathione is reduced to GSH in the presence of NADPH (131). Therefore, UCA1 regulates GSH and
NADPH expression, thereby enhancing the antioxidant effects of GPX4
and inhibiting ferroptosis in tumor cells by decreasing ROS
production (132). The
transcription factor NRF2 also has antioxidant properties and can
activate downstream antioxidant factors. NRF2 can promote the
production of NADPH via the pentose phosphate pathway, thereby
enhancing antioxidant activity. Additionally, NRF2 activates
transcription of GSH and GPX family genes, while GPX4 is activated
to exert antioxidant effects during ferroptosis (133). The balance of intracellular iron
levels is key for cell survival. The co-function of transferrin and
transferrin receptor is required for the absorption of
Fe3+ by cells. Subsequently, Fe3+ becomes
Fe2+ through a redox reaction and it is then stored in
the iron pool (134). Ferroptosis
occurs when Fe2+ donates electrons to oxygen in the
Fenton reaction to form ROS, which catalyzes generation of lipid
free oxygen radicals (135). In
HCC, for example, expression of the ncRNA plasmacytoma variant
translocation 1 (PVT1) is increased; PVT1 binds to miR-150 and
regulates the target hypoxia-inducible gene (HIG2) (136). HIG2 is involved in ferroptosis
and iron uptake via hypoxia-induced protein expression. Decreased
transferrin receptor and increased ferritin light chain expression
are observed when PVT1 is silenced, thereby decreasing iron uptake
and affecting ferroptosis (137).
The induction of ferroptosis by erastin results in upregulation of
the lncRNA GA binding protein subunit β1 (GABPB1)-antisense RNA 1,
blocking of the transcription and translation of GABPB1 and the
inhibition of peroxidase 5 (PRDX5) expression. PRDX5 is less
effective than non tumor tissue at preventing ROS production from
H2O2, which ultimately impairs the
antioxidant capacity of tumor cells, resulting in increased cell
death (138). Melanoma
glucose-6-phosphate dehydrogenase (G6PD) may stimulate NADPH
production, thereby activating NADPH oxidase 4 (NOX4) (139). Superoxide and ROS are produced by
NOX4, a transmembrane protein. When growth arrest specific 5 is
silenced, G6PD and NOX4 activity is increased (140). Furthermore, ferroptosis is
induced by G6PD and NOX4 in tumor cells (141). Inhibition of iron-induced cell
death by targeting the miR-106B-5p/acyl-coenzyme A synthetase long
chain family member 4 axis is a mechanism by which H19 promotes
cancer progression (142).
Iron-induced death is also regulated by PVT1 via activation of TFR1
and TP53 by miR-214. The PVT1/miR-214-3p/GPX4 signaling pathway
activates ketamine and inhibits the proliferation of HCC cells both
in vitro and in vivo, leading to iron-induced cell
death (143). In addition, lncRNA
ZNFX antisense RNA 1 acts as a competing endogenous RNA for
iron-induced death via the miR-150-5p/SLC38A1 axis (144). Multiple other lncRNAs are
involved in iron-induced tumor cell death. Further studies are
needed to investigate the specific mechanism by which lncRNA
regulates iron-related cell death.
circRNAs are formed by the 3′ and 5′ ends of mRNAs.
According to their structural features, they are primarily
classified into three categories, namely, exonic, circular intronic
and exonic intronic circRNAs, and they function as miRNA sponges
and protein scaffolds (145).
Compared with miRNAs, the abundance, conservation and tissue
specificity of circRNAs make them better biomarkers for certain
pathological states. For example, circrNA-002178 is abnormally
expressed in lung cancer, which is of great help in the diagnosis
of lung cancer (146). The
majority of circRNAs are highly stable and specific. Cancer cells
express abnormal levels of circRNAs, which are involved in cell
proliferation, autophagy and apoptosis, among other aspects of
programmed cell death (147).
circRNAs affect the post-transcriptional stability of miRNAs by
altering their target gene expression. In tumor cells, circRNAs
serve as sponges and regulate ferroptosis (148). Furthermore, they enhance tumor
cell proliferation and invasion by sponging miR-761 and activating
integrin β8 (ITGB8) in glioma, inhibit ferroptosis by activating
ITGB8 and inhibit apoptosis by targeting MET, while knockdown of
circ-tubulin kinase 2 promotes erastin-induced ferroptosis
(149). In addition, 526 circRNAs
are dysregulated in cervical cancer cells according to a previous
high-throughput microarray-based study (150). circRNAs are predominantly
involved in GSH metabolism according to a previous bioinformatics
analysis (151). However,
associated downstream factors and miRNAs have not been screened
(152). Therefore, further
studies on regulation of circRNAs in ferroptosis are needed
(Fig. 2).
Chemical modifications of the tails of four core
histone proteins (H2A, H2B, H3 and H4) alter the interactions
between histones and other nuclear proteins, according to a recent
study (153). Consequently,
histone modification modifies expression of target genes. For
example, the ferroptosis-associated genes GPX4 and SLC7A11 are
regulated by histone modifications in non-Hodgkin lymphoma
(154,155). Therefore, regulation of
ferroptosis is dependent on histone modification.
The histone H3 lysine 9 demethylase that regulates
SLC7A11 expression in response to erastin-induced ferroptosis is
lysine demethylase 3B (156). E1A
binding protein p300-CREB binding protein (CREBBP), which is
involved in transactivation of genes, acetylates nuclear factor
erythroid-derived 2-like 2 (NFE2L2) (157). Mouse double minute 2 homolog is a
proto-oncogene associated with TP53 stability and activation, while
cyclin-dependent kinase inhibitor 2A/ARF promotes its degradation.
Ferroptosis is promoted by ARF independently of TP53 (158). Cancer is frequently caused by
TP53 inactivation or mutations. A number of genes controlled by
TP53 serve key roles in various cellular processes such as cell
proliferation, cell cycle progression, cell death and response to
DNA damage. Cancer cells, when activated by TP53, not only promote
tumor growth but also inhibit ferroptosis (159). ARF protein disrupts CREBBP-NFE2L2
interaction, resulting in attenuated acetylation of NFE2L2 and the
decreased expression of SLC7A11 mediated by NFE2L2 (160). Furthermore, members of the
bromodomain (BRD) family are epigenetic regulators that recognize
acetylated lysine residues on histones. JQ1, an inhibitor of the
BRD4 complex, induces ferroptosis in breast and lung cancer cells
by decreasing expression of GPX4, SLC7A11 and SLC3A2. Other
anti-ferroptotic genes are also regulated by BRD4 (161).
Cancer cells overexpress deubiquitinase OUT
deubiquitinase ubiquitin aldehyde binding 1 (OTUB1), which
stabilizes cystine transporter SLC7A11, inhibits ferroptosis and
promotes tumor growth (162).
BRCA1-associated protein 1 (BAP1) decreases SLC7A11 promoter H2A
ubiquitination via an H2A deubiquitinase that functions as a tumor
suppressor. As a result, ferroptosis is regulated and SLC7A11
expression is inhibited. The H2A ubiquitin ligases BAP1 and protein
regulator of cytokinesis 1 inhibit expression of SLC7A11. The tumor
suppressor deubiquitinating enzymes (DUB) inactivates P53 and BAP1
by downregulating SLC7A11 expression or enhancing OTUB1 or CD44
expression to stabilize SLC7A11 (153). As a result, tumor growth is
suppressed and proteasomal activity and apoptosis are inhibited.
The DUB inhibitor ubiquitin-specific protease (USP7) enhances
caspase-dependent apoptosis during ferroptosis while degrading
GPX4, which results in ubiquitinated protein accumulation and cell
death (163). The expression of
H2Bub1, which binds to the regulatory region of SLC7A11, is
decreased by P53, thereby inhibiting SLC7A11 expression (164). Furthermore, ferritin degradation
occurs via lysosomal or proteasomal mechanisms. Via ferritin heavy
chain 1, NCOA4 binds to iron-rich ferritin on autophagosomes, thus
transporting it to lysosomes for iron release. The ubiquitin ligase
HERC2, which affects stability of proteins, ubiquitinates and
degrades NCOA4 in environments with high iron concentrations.
Therefore, ferroptosis is suppressed by inhibition of NCOA4
(165).
The four aforementioned histones are acetylated on
specific lysine residues, which is associated not only with gene
transcription but also with DNA replication and repair (166). During transcription, histone
acetylation induces HAT to acetylate lysine residues in histones,
while histone deacetylation inhibits HDAC (167). HAT and HDAC catalyze histone
acetylation and deacetylation, respectively (168). Patients with cancer have a poor
prognosis when GPX4 expression is higher in tumor than in normal
tissue (169). In tumor cells,
high GPX4 levels may be due to epigenetic regulation such as
methylation of DNA and histones and acetylation of histones, as
reported in a previous study that analyzed upstream regulation of
GPX4 (170). Mutant P533KR causes
lipid peroxidation and ferroptosis via decreased acetylation and
cysteine uptake (171). As a
result of hydrolysis of cystinase, GSH is synthesized and GSH
metabolism is affected. Cysteinase depletion also cause ferroptosis
(Table I) (172).
Iron is a key element and its absorption, transport,
storage and excretion are tightly regulated in the human body. Iron
promotes formation of free radicals; excessive free radicals can be
harm to the body by promoting tumor formation and subsequent
metastasis. Study of iron metabolism in tumors has demonstrated
that the combined actions of iron transporters and iron efflux
regulator hepcidin promote tumorigenesis (173). Ferroptosis is an iron-dependent
mechanism involving production of ROS. Mechanisms controlling
transcripts that encode iron-regulated proteins, known as
epigenetic regulatory mechanisms, have been reported in tumor cells
(57). Furthermore, epigenetic
changes in gene expression levels are not induced by changes in
genomic sequences but DNA methylation, chromatin histone
modifications and ncRNA regulation. Thus, abnormal epigenetic
mechanisms affect gene transcription, which promotes tumor
occurrence and development with certain generality and tissue
specificity (174). Epigenetic
modifications are key for tumor cell death and ferroptosis. The
aforementioned findings may provide new therapeutic insights for
the treatment of cancer in the future.
The present review summarizes the research of
epigenetic inheritance and ferroptosis. The three main regulatory
mechanisms of ferroptosis and ROS were introduced, with emphasis on
the association between ROS and ferroptosis and epigenetic
inheritance. The process of iron homeostasis metabolism and the
association between iron homeostasis and reactive oxygen species
were also described, which has not performed in the other two
papers (175,176). In addition, the epigenetic
regulation mechanism of ferroptosis was introduced, especially the
association between ncRNA, miRNA and circRNA in ferroptosis and
histone ubiquitination, methylation and acetylation with
ferroptosis. The aforementioned findings may provide novel options
for the treatment of tumors in terms of epigenetic regulation of
ferroptosis (177).
Not applicable.
Funding: No funding was received.
Not applicable.
JWu, SZ and PW wrote the manuscript. JWa, JH, TW
and LG constructed figures and tables. DL, QM and HP performed the
literature review. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Subramanian S, Geng H and Tan XD: Cell
death of intestinal epithelial cells in intestinal diseases. Sheng
Li Xue Bao. 72:308–324. 2020.PubMed/NCBI
|
|
3
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Su LJ, Zhang JH, Gomez H, Murugan R, Hong
X, Xu D, Jiang F and Peng ZY: Reactive oxygen species-induced lipid
peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med
Cell Longev. 2019:50808432019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
D'Arcy MS: Cell death: A review of the
major forms of apoptosis, necrosis and autophagy. Cell Biol Int.
43:582–592. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vanden Berghe T, Linkermann A,
Jouan-Lanhouet S, Walczak H and Vandenabeele P: Regulated necrosis:
The expanding network of non-apoptotic cell death pathways. Nat Rev
Mol Cell Biol. 15:135–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hirschhorn T and Stockwell BR: The
development of the concept of ferroptosis. Free Radic Biol Med.
133:130–143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang X, Sui S, Wang L, Li H, Zhang L, Xu
S and Zheng X: Inhibition of tumor propellant glutathione
peroxidase 4 induces ferroptosis in cancer cells and enhances
anticancer effect of cisplatin. J Cell Physiol. 235:3425–3437.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deans C and Maggert KA: What do you mean,
‘epigenetic’? Genetics. 199:887–896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu J, Xiong Y, Zhang Y, Wen J, Cai N,
Cheng K, Liang H and Zhang W: The molecular mechanisms of
regulating oxidative stress-induced ferroptosis and therapeutic
strategy in tumors. Oxid Med Cell Longev. 2020:88107852020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei Y, Lv H, Shaikh AB, Han W, Hou H,
Zhang Z, Wang S and Shang P: Directly targeting glutathione
peroxidase 4 may be more effective than disrupting glutathione on
ferroptosis-based cancer therapy. Biochim Biophys Acta Gen Subj.
1864:1295392020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang X, Wang H, Han D, Xie E, Yang X, Wei
J, Gu S, Gao F, Zhu N, Yin X, et al: Ferroptosis as a target for
protection against cardiomyopathy. Proc Natl Acad Sci USA.
116:2672–2680. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bogdan AR, Miyazawa M, Hashimoto K and
Tsuji Y: Regulators of iron homeostasis: New players in metabolism,
cell death, and disease. Trends Biochem Sci. 41:274–286. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anderson CP, Shen M, Eisenstein RS and
Leibold EA: Mammalian iron metabolism and its control by iron
regulatory proteins. Biochim Biophys Acta. 1823:1468–1483. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kato J, Kobune M, Ohkubo S, Fujikawa K,
Tanaka M, Takimoto R, Takada K, Takahari D, Kawano Y, Kohgo Y and
Niitsu Y: Iron/IRP-1-dependent regulation of mRNA expression for
transferrin receptor, DMT1 and ferritin during human erythroid
differentiation. Exp Hematol. 35:879–887. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bolotta A, Abruzzo PM, Baldassarro VA,
Ghezzo A, Scotlandi K, Marini M and Zucchini C: New insights into
the hepcidin-ferroportin axis and iron homeostasis in iPSC-derived
cardiomyocytes from friedreich's ataxia patient. Oxid Med Cell
Longev. 2019:76230232019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao G, Li J, Zhang Y and Chang YZ:
Cellular iron metabolism and regulation. Adv Exp Med Biol.
1173:21–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arruda SF, Ramos LV, Barbosa JLA, Hankins
NAC, Rodrigues PAM and Cunha MSBD: The action of JAK/STAT3 and
BMP/HJV/SMAD signaling pathways on hepcidin suppression by
tucum-do-cerrado in a normal and iron-enriched diets. Nutrients.
12:15152020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Babitt JL, Huang FW, Wrighting DM, Xia Y,
Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, et
al: Bone morphogenetic protein signaling by hemojuvelin regulates
hepcidin expression. Nat Genet. 38:531–539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu PB, Hong CC, Sachidanandan C, Babitt
JL, Deng DY, Hoyng SA, Lin HY, Bloch KD and Peterson RT:
Dorsomorphin inhibits BMP signals required for embryogenesis and
iron metabolism. Nat Chem Biol. 4:33–41. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Truksa J, Lee P and Beutler E: Two BMP
responsive elements, STAT, and bZIP/HNF4/COUP motifs of the
hepcidin promoter are critical for BMP, SMAD1, and HJV
responsiveness. Blood. 113:688–695. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Srole DN and Ganz T: Erythroferrone
structure, function, and physiology: Iron homeostasis and beyond. J
Cell Physiol. 236:4888–4901. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Duan L, Yin X, Meng H, Fang X, Min J and
Wang F: Progress on epigenetic regulation of iron homeostasis.
Zhejiang Da Xue Xue Bao Yi Xue Ban. 49:58–70. 2020.(In Chinese).
PubMed/NCBI
|
|
24
|
Patnaik MM and Tefferi A: Myelodysplastic
syndromes with ring sideroblasts (MDS-RS) and
MDS/myeloproliferative neoplasm with RS and thrombocytosis
(MDS/MPN-RS-T)-‘2021 update on diagnosis, risk-stratification, and
management’. Am J Hematol. 96:379–394. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rawat PS, Jaiswal A, Khurana A, Bhatti JS
and Navik U: Doxorubicin-induced cardiotoxicity: An update on the
molecular mechanism and novel therapeutic strategies for effective
management. Biomed Pharmacother. 139:1117082021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blackledge NP and Klose R: CpG island
chromatin: A platform for gene regulation. Epigenetics. 6:147–152.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Illingworth RS, Gruenewald-Schneider U,
Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews
R and Bird AP: Orphan CpG islands identify numerous conserved
promoters in the mammalian genome. PLoS Genet. 6:e10011342010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Grand RS, Burger L, Gräwe C, Michael AK,
Isbel L, Hess D, Hoerner L, Iesmantavicius V, Durdu S, Pregnolato
M, et al: BANP opens chromatin and activates CpG-island-regulated
genes. Nature. 596:133–137. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maunakea AK, Nagarajan RP, Bilenky M,
Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C,
Zhao Y, et al: Conserved role of intragenic DNA methylation in
regulating alternative promoters. Nature. 466:253–257. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Horvath S and Raj K: DNA methylation-based
biomarkers and the epigenetic clock theory of ageing. Nat Rev
Genet. 19:371–384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye Q, Trivedi M, Zhang Y, Böhlke M,
Alsulimani H, Chang J, Maher T, Deth R and Kim J: Brain iron
loading impairs DNA methylation and alters GABAergic function in
mice. FASEB J. 33:2460–2471. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Macková E, Hrušková K, Bendová P, Vávrová
A, Jansová H, Hašková P, Kovaříková P, Vávrová K and Simůnek T:
Methyl and ethyl ketone analogs of salicylaldehyde isonicotinoyl
hydrazone: Novel iron chelators with selective antiproliferative
action. Chem Biol Interact. 197:69–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang WC, Lin SF, Wang SC, Tsai WC, Wu CC
and Wu SC: The effects of human BDH2 on the cell cycle,
differentiation, and apoptosis and associations with leukemia
transformation in myelodysplastic syndrome. Int J Mol Sci.
21:30332020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma S, Henson ES, Chen Y and Gibson SB:
Ferroptosis is induced following siramesine and lapatinib treatment
of breast cancer cells. Cell Death Dis. 7:e23072016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kajarabille N and Latunde-Dada GO:
Programmed cell-death by ferroptosis: Antioxidants as mitigators.
Int J Mol Sci. 20:49682019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao J, Luo T and Wang J: Gene
interfered-ferroptosis therapy for cancers. Nat Commun.
12:53112021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang F, Wang J, Shen Y, Li H, Rausch WD
and Huang X: Iron dyshomeostasis and ferroptosis: A new Alzheimer's
disease hypothesis? Front Aging Neurosci. 14:8305692022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Y, Zhang S, Wang X, Guo W, Wang L,
Zhang D, Yuan L, Zhang Z, Xu Y and Liu S: Disordered signaling
governing ferroportin transcription favors breast cancer growth.
Cell Signal. 27:168–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Udali S, Castagna A, Corbella M,
Ruzzenente A, Moruzzi S, Mazzi F, Campagnaro T, De Santis D,
Franceschi A, Pattini P, et al: Hepcidin and DNA promoter
methylation in hepatocellular carcinoma. Eur J Clin Invest.
48:e128702018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Al-Amer O and Alsharif KF: Frequency of
the HAMP (c.-582 A>G) polymorphism in iron deficiency in Saudi
Arabia. Pak J Biol Sci. 24:146–150. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sies H: Oxidative stress: A concept in
redox biology and medicine. Redox Biol. 4:180–183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang Z, Liu Y, He M and Bu W: Chemodynamic
therapy: Tumour microenvironment-mediated fenton and fenton-like
reactions. Angew Chem Int Ed Engl. 58:946–956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Miller CJ, Rose AL and Waite TD:
Importance of iron complexation for fenton-mediated hydroxyl
radical production at circumneutral pH. Front Mar Sci. 3:1342016.
View Article : Google Scholar
|
|
44
|
Yu L, Lin Z, Cheng X, Chu J, Li X, Chen C,
Zhu T, Li W, Lin W and Tang W: Thorium inhibits human respiratory
chain complex IV (cytochrome c oxidase). J Hazard Mater.
424:1275462022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rattanawong K, Koiso N, Toda E, Kinoshita
A, Tanaka M, Tsuji H and Okamoto T: Regulatory functions of ROS
dynamics via glutathione metabolism and glutathione peroxidase
activity in developing rice zygote. Plant J. 108:1097–1115. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peoples JN, Saraf A, Ghazal N, Pham TT and
Kwong JQ: Mitochondrial dysfunction and oxidative stress in heart
disease. Exp Mol Med. 51:1–13. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Prasad S, Gupta SC and Tyagi AK: Reactive
oxygen species (ROS) and cancer: Role of antioxidative
nutraceuticals. Cancer Lett. 387:95–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang R, Kang KA, Kim KC, Na SY, Chang WY,
Kim GY, Kim HS and Hyun JW: Oxidative stress causes epigenetic
alteration of CDX1 expression in colorectal cancer cells. Gene.
524:214–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang Y, Karakhanova S, Hartwig W, D'Haese
JG, Philippov PP, Werner J and Bazhin AV: Mitochondria and
mitochondrial ROS in cancer: Novel targets for anticancer therapy.
J Cell Physiol. 231:2570–2581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Islam MT: Oxidative stress and
mitochondrial dysfunction-linked neurodegenerative disorders.
Neurol Res. 39:73–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao Y, Zhang J, Zheng Y, Zhang Y, Zhang
XJ, Wang H, Du Y, Guan J, Wang X and Fu J: NAD+ improves
cognitive function and reduces neuroinflammation by ameliorating
mitochondrial damage and decreasing ROS production in chronic
cerebral hypoperfusion models through Sirt1/PGC-1α pathway. J
Neuroinflammation. 18:2072021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Uittenbogaard M, Baxter KK and Chiaramello
A: The neurogenic basic helix-loop-helix transcription factor
NeuroD6 confers tolerance to oxidative stress by triggering an
antioxidant response and sustaining the mitochondrial biomass. ASN
Neuro. 2:e000342010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hou K, Chen Y, Zhu D, Chen G, Chen F, Xu
N, Barakat K, Zheng J, Xie X and Chen R: Curcumin inhibits high
glucose oxidative stress and apoptosis in pancreatic beta cells via
CHOP/PCG-1a and pERK1/2. Front Biosci (Landmark Ed). 25:1974–1984.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tang G, Li S, Zhang C, Chen H, Wang N and
Feng Y: Clinical efficacies, underlying mechanisms and molecular
targets of Chinese medicines for diabetic nephropathy treatment and
management. Acta Pharm Sin B. 11:2749–2767. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Su X, Chu Y, Kordower JH, Li B, Cao H,
Huang L, Nishida M, Song L, Wang D and Federoff HJ: PGC-1α promoter
methylation in Parkinson's disease. PLoS One. 10:e01340872015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Y, Chen M, Xu Y, Yu X, Xiong T, Du M,
Sun J, Liu L, Tang Y and Yao P: Iron-mediated lysosomal membrane
permeabilization in ethanol-induced hepatic oxidative damage and
apoptosis: Protective effects of quercetin. Oxid Med Cell Longev.
2016:41476102016.PubMed/NCBI
|
|
57
|
Park E and Chung SW: ROS-mediated
autophagy increases intracellular iron levels and ferroptosis by
ferritin and transferrin receptor regulation. Cell Death Dis.
10:8222019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen H, Xu C, Yu Q, Zhong C, Peng Y, Chen
J and Chen G: Comprehensive landscape of STEAP family functions and
prognostic prediction value in glioblastoma. J Cell Physiol.
236:2988–3000. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Song Q, Peng S, Sun Z, Heng X and Zhu X:
Temozolomide drives ferroptosis via a DMT1-dependent pathway in
glioblastoma cells. Yonsei Med J. 62:843–849. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cheung EC and Vousden KH: The role of ROS
in tumour development and progression. Nat Rev Cancer. 22:280–297.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wei S, Qiu T, Yao X, Wang N, Jiang L, Jia
X, Tao Y, Wang Z, Pei P, Zhang J, et al: Arsenic induces pancreatic
dysfunction and ferroptosis via mitochondrial
ROS-autophagy-lysosomal pathway. J Hazard Mater. 384:1213902020.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou B, Liu J, Kang R, Klionsky DJ,
Kroemer G and Tang D: Ferroptosis is a type of autophagy-dependent
cell death. Semin Cancer Biol. 66:89–100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cao JY and Dixon SJ: Mechanisms of
ferroptosis. Cell Mol Life Sci. 73:2195–2209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhuang Y, Li T, Xiao H, Wu J, Su S, Dong
X, Hu X, Hua Q, Liu J, Shang W, et al: LncRNA-H19 drives
cardiomyocyte senescence by targeting miR-19a/socs1/p53 axis. Front
Pharmacol. 12:6318352021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C
and Li B: Ferroptosis, a new form of cell death: Opportunities and
challenges in cancer. J Hematol Oncol. 12:342019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Koppula P, Zhuang L and Gan B: Cystine
transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient
dependency, and cancer therapy. Protein Cell. 12:599–620. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang H, He Y, Wang JX, Chen MH, Xu JJ,
Jiang MH, Feng YL and Gu YF: miR-30-5p-mediated ferroptosis of
trophoblasts is implicated in the pathogenesis of preeclampsia.
Redox Biol. 29:1014022020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu MR, Zhu WT and Pei DS: System
Xc−: A key regulatory target of ferroptosis in cancer.
Invest New Drugs. 39:1123–1131. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang L, Liu Y, Du T, Yang H, Lei L, Guo M,
Ding HF, Zhang J, Wang H, Chen X and Yan C: ATF3 promotes
erastin-induced ferroptosis by suppressing system Xc. Cell Death
Differ. 27:662–675. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kong R, Wang N, Han W, Bao W and Lu J:
IFNγ-mediated repression of system xc− drives
vulnerability to induced ferroptosis in hepatocellular carcinoma
cells. J Leukoc Biol. 110:301–314. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Floros KV, Cai J, Jacob S, Kurupi R,
Fairchild CK, Shende M, Coon CM, Powell KM, Belvin BR, Hu B, et al:
MYCN-amplified neuroblastoma is addicted to iron and vulnerable to
inhibition of the system Xc-/glutathione axis. Cancer Res.
81:1896–1908. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ding Y, Chen X, Liu C, Ge W, Wang Q, Hao
X, Wang M, Chen Y and Zhang Q: Identification of a small molecule
as inducer of ferroptosis and apoptosis through ubiquitination of
GPX4 in triple negative breast cancer cells. J Hematol Oncol.
14:192021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Imai H, Matsuoka M, Kumagai T, Sakamoto T
and Koumura T: Lipid peroxidation-dependent cell death regulated by
GPx4 and ferroptosis. Curr Top Microbiol Immunol. 403:143–170.
2017.PubMed/NCBI
|
|
74
|
Li H, Liu W, Zhang X, Wu F, Sun D and Wang
Z: Ketamine suppresses proliferation and induces ferroptosis and
apoptosis of breast cancer cells by targeting KAT5/GPX4 axis.
Biochem Biophys Res Commun. 585:111–116. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Tang X, Ding H, Liang M, Chen X, Yan Y,
Wan N, Chen Q, Zhang J and Cao J: Curcumin induces ferroptosis in
non-small-cell lung cancer via activating autophagy. Thorac Cancer.
12:1219–1230. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang Y, Kong Y, Ma Y, Ni S, Wikerholmen
T, Xi K, Zhao F, Zhao Z, Wang J, Huang B, et al: Loss of COPZ1
induces NCOA4 mediated autophagy and ferroptosis in glioblastoma
cell lines. Oncogene. 40:1425–1439. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bersuker K, Hendricks JM, Li Z, Magtanong
L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al:
The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit
ferroptosis. Nature. 575:688–692. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang CX, Chen LH, Zhuang HB, Shi ZS, Chen
ZC, Pan JP and Hong ZS: Auriculasin enhances ROS generation to
regulate colorectal cancer cell apoptosis, ferroptosis, oxeiptosis,
invasion and colony formation. Biochem Biophys Res Commun.
587:99–106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Santoro MM: The antioxidant role of
non-mitochondrial CoQ10: mystery solved! Cell Metab. 31:13–15.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Doll S, Freitas FP, Shah R, Aldrovandi M,
da Silva MC, Ingold I, Goya Grocin A, Xavier da Silva TN, Panzilius
E, Scheel CH, et al: FSP1 is a glutathione-independent ferroptosis
suppressor. Nature. 575:693–698. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zheng J and Conrad M: The metabolic
underpinnings of ferroptosis. Cell Metab. 32:920–937. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hu Q, Wei W, Wu D, Huang F, Li M, Li W,
Yin J, Peng Y, Lu Y, Zhao Q and Liu L: Blockade of GCH1/BH4 axis
activates ferritinophagy to mitigate the resistance of colorectal
cancer to erastin-induced ferroptosis. Front Cell Dev Biol.
10:8103272022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Qiu C, Liu T, Luo D, Luan D, Cheng L and
Wang S: Novel therapeutic savior for osteosarcoma: The endorsement
of ferroptosis. Front Oncol. 12:7460302022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Gao M, Fan K, Chen Y, Zhang G, Chen J and
Zhang Y: Understanding the mechanistic regulation of ferroptosis in
cancer: The gene matters. J Genet Genomics. S1673-8527(22)00160-6.
2022.(Epub ahead of print). View Article : Google Scholar
|
|
85
|
Kraft VAN, Bezjian CT, Pfeiffer S,
Ringelstetter L, Müller C, Zandkarimi F, Merl-Pham J, Bao X,
Anastasov N, Kössl J, et al: GTP cyclohydrolase
1/tetrahydrobiopterin counteract ferroptosis through lipid
remodeling. ACS Cent Sci. 6:41–53. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chew GT and Watts GF: Coenzyme Q10 and
diabetic endotheliopathy: Oxidative stress and the ‘recoupling
hypothesis’. QJM. 97:537–548. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ilango S, Paital B, Jayachandran P, Padma
PR and Nirmaladevi R: Epigenetic alterations in cancer. Front
Biosci (Landmark Ed). 25:1058–1109. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wu SC and Zhang Y: Active DNA
demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol.
11:607–620. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Goll MG and Bestor TH: Eukaryotic cytosine
methyltransferases. Annu Rev Biochem. 74:481–514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lu F, Liu Y, Jiang L, Yamaguchi S and
Zhang Y: Role of Tet proteins in enhancer activity and telomere
elongation. Genes Dev. 28:2103–2119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jiang J, Yan T and Guo F: Global DNA 5hmC
and CK195hmC+ contents: A promising biomarker for
predicting prognosis in small hepatocellular carcinoma. Curr Oncol.
28:3758–3770. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ito S, Shen L, Dai Q, Wu SC, Collins LB,
Swenberg JA, He C and Zhang Y: Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Science. 333:1300–1303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hashimoto H, Vertino PM and Cheng X:
Molecular coupling of DNA methylation and histone methylation.
Epigenomics. 2:657–669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jara-Espejo M and Line SR: DNA
G-quadruplex stability, position and chromatin accessibility are
associated with CpG island methylation. FEBS J. 287:483–495. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Horii T and Hatada I: Regulation of CpG
methylation by Dnmt and Tet in pluripotent stem cells. J Reprod
Dev. 62:331–335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Van Tongelen A, Loriot A and De Smet C:
Oncogenic roles of DNA hypomethylation through the activation of
cancer-germline genes. Cancer Lett. 396:130–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Jair KW, Bachman KE, Suzuki H, Ting AH,
Rhee I, Yen RW, Baylin SB and Schuebel KE: De novo CpG island
methylation in human cancer cells. Cancer Res. 66:682–692. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Mendoza-Pérez J, Gu J, Herrera LA, Tannir
NM, Matin SF, Karam JA, Huang M, Chang DW, Wood CG and Wu X:
Genomic DNA hypomethylation and risk of renal cell carcinoma: A
case-control study. Clin Cancer Res. 22:2074–2082. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhang L, Xiao X and Xu ZC: iPromoter-5mC:
A novel fusion decision predictor for the identification of
5-methylcytosine sites in genome-wide DNA promoters. Front Cell Dev
Biol. 8:6142020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Robert MF, Morin S, Beaulieu N, Gauthier
F, Chute IC, Barsalou A and MacLeod AR: DNMT1 is required to
maintain CpG methylation and aberrant gene silencing in human
cancer cells. Nat Genet. 33:61–65. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Choi SJ, Jung SW, Huh S, Chung YS, Cho H
and Kang H: Alteration of DNA methylation in gastric cancer with
chemotherapy. J Microbiol Biotechnol. 27:1367–1378. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Logie E, Van Puyvelde B, Cuypers B,
Schepers A, Berghmans H, Verdonck J, Laukens K, Godderis L,
Dhaenens M, Deforce D and Vanden Berghe W: Ferroptosis induction in
multiple myeloma cells triggers DNA methylation and histone
modification changes associated with cellular senescence. Int J Mol
Sci. 22:122342021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xu Y, Hong M, Kong D, Deng J, Zhong Z and
Liang J: Ferroptosis-associated DNA methylation signature predicts
overall survival in patients with head and neck squamous cell
carcinoma. BMC Genomics. 23:632022. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang
Y, Shi Y, Shen Y, Liu X, Lai W, et al: A G3BP1-interacting lncRNA
promotes ferroptosis and apoptosis in cancer via nuclear
sequestration of p53. Cancer Res. 78:3484–3496. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu
X, Lai W, Liu Y, Wang X, Xiao D, et al: EGLN1/c-Myc induced
lymphoid-specific helicase inhibits ferroptosis through lipid
metabolic gene expression changes. Theranostics. 7:3293–3305. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zhang X, Huang Z, Xie Z, Chen Y, Zheng Z,
Wei X, Huang B, Shan Z, Liu J, Fan S, et al: Homocysteine induces
oxidative stress and ferroptosis of nucleus pulposus via enhancing
methylation of GPX4. Free Radic Biol Med. 160:552–565. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
van Roy F: Beyond E-cadherin: Roles of
other cadherin superfamily members in cancer. Nat Rev Cancer.
14:121–134. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Tan L and Shi YG: Tet family proteins and
5-hydroxymethylcytosine in development and disease. Development.
139:1895–1902. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Tang Y, Li C, Zhang YJ and Wu ZH:
Ferroptosis-related long non-coding RNA signature predicts the
prognosis of Head and neck squamous cell carcinoma. Int J Biol Sci.
17:702–711. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wang J, Zhu S, Meng N, He Y, Lu R and Yan
GR: ncRNA-encoded peptides or proteins and cancer. Mol Ther.
27:1718–1725. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhang H, Deng T, Liu R, Ning T, Yang H,
Liu D, Zhang Q, Lin D, Ge S, Bai M, et al: CAF secreted miR-522
suppresses ferroptosis and promotes acquired chemo-resistance in
gastric cancer. Mol Cancer. 19:432020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hill M and Tran N: miRNA interplay:
Mechanisms and consequences in cancer. Dis Model Mech.
14:dmm0476622021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Mishra S, Yadav T and Rani V: Exploring
miRNA based approaches in cancer diagnostics and therapeutics. Crit
Rev Oncol Hematol. 98:12–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sengupta D, Deb M, Kar S, Pradhan N,
Parbin S, Kirtana R, Singh SP, Suma SG, Niharika, Roy A, et al:
Dissecting miRNA facilitated physiology and function in human
breast cancer for therapeutic intervention. Semin Cancer Biol.
72:46–64. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Lande K, Gupta J, Ranjan R, Kiran M,
Torres Solis LF, Solís Herrera A, Aliev G and Karnati R: Exosomes:
Insights from retinoblastoma and other eye cancers. Int J Mol Sci.
21:70552020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Luo M, Wu L, Zhang K, Wang H, Zhang T,
Gutierrez L, O'Connell D, Zhang P, Li Y, Gao T, et al: miR-137
regulates ferroptosis by targeting glutamine transporter SLC1A5 in
melanoma. Cell Death Differ. 25:1457–1472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Tomita K, Nagasawa T, Kuwahara Y, Torii S,
Igarashi K, Roudkenar MH, Roushandeh AM, Kurimasa A and Sato T:
MiR-7-5p is involved in ferroptosis signaling and radioresistance
thru the generation of ROS in radioresistant HeLa and SAS cell
lines. Int J Mol Sci. 22:83002021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Chen D, Fan Z, Rauh M, Buchfelder M,
Eyupoglu IY and Savaskan N: ATF4 promotes angiogenesis and neuronal
cell death and confers ferroptosis in a xCT-dependent manner.
Oncogene. 36:5593–5608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Gomaa A, Peng D, Chen Z, Soutto M,
Abouelezz K, Corvalan A and El-Rifai W: Epigenetic regulation of
AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci Rep.
9:169702019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yu W, Yao J, Lyu P, Zhou J, Chen X, Liu X
and Xiao S: XPG is modulated by miR-4715-3p and rs873601 genotypes
in lung cancer. Cancer Manag Res. 13:3417–3427. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhuang ST, Cai YJ, Liu HP, Qin Y and Wen
JF: LncRNA NEAT1/miR-185-5p/IGF2 axis regulates the invasion and
migration of colon cancer. Mol Genet Genomic Med. 8:e11252020.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Marengo B, Pulliero A, Izzotti A and
Domenicotti C: miRNA regulation of glutathione homeostasis in
cancer initiation, progression and therapy resistance. Microrna.
9:187–197. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Hu X, Miao J, Zhang M, Wang X, Wang Z, Han
J, Tong D and Huang C: miRNA-103a-3p promotes human gastric cancer
cell proliferation by targeting and suppressing ATF7 in vitro. Mol
Cells. 41:390–400. 2018.PubMed/NCBI
|
|
126
|
Zhi Y, Gao L, Wang B, Ren W, Liang KX and
Zhi K: Ferroptosis holds novel promise in treatment of cancer
mediated by non-coding RNAs. Front Cell Dev Biol. 9:6869062021.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Bridges MC, Daulagala AC and Kourtidis A:
LNCcation: lncRNA localization and function. J Cell Biol.
220:e2020090452021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu
N, Shi Y, Chen L, Xiao D, Yu F, et al: Long noncoding RNA LINC00336
inhibits ferroptosis in lung cancer by functioning as a competing
endogenous RNA. Cell Death Differ. 26:2329–2343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Wang Y, Li X, Chen W and Wu W: The common
region of lncRNAs UCA1 and UCA1α contributes to the bladder cancer
tumorigenesis. Eur J Cancer Prev. 30:389–392. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Kim S, Kim JE, Kim YH, Hwang T, Kim SK, Xu
WJ, Shin JY, Kim JI, Choi H, Kim HC, et al: Glutaminase 2
expression is associated with regional heterogeneity of
5-aminolevulinic acid fluorescence in glioblastoma. Sci Rep.
7:122212017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Chen J, Hu Q, Zhang BF, Liu XP, Yang S and
Jiang H: Long noncoding RNA UCA1 inhibits ischaemia/reperfusion
injury induced cardiomyocytes apoptosis via suppression of
endoplasmic reticulum stress. Genes Genomics. 41:803–810. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Shin D, Kim EH, Lee J and Roh JL: Nrf2
inhibition reverses resistance to GPX4 inhibitor-induced
ferroptosis in head and neck cancer. Free Radic Biol Med.
129:454–462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Yang Y, Tian Q, Wu S, Li Y, Yang K, Yan Y,
Shang L, Li A and Zhang L: Blue light-triggered
Fe2+-release from monodispersed ferrihydrite
nanoparticles for cancer iron therapy. Biomaterials.
271:1207392021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Xie Z, Hou H, Luo D, An R, Zhao Y and Qiu
C: ROS-dependent lipid peroxidation and reliant antioxidant
ferroptosis-suppressor-protein 1 in rheumatoid arthritis: A covert
clue for potential therapy. Inflammation. 44:35–47. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Xu Y, Luo X, He W, Chen G, Li Y, Li W,
Wang X, Lai Y and Ye Y: Long non-coding RNA PVT1/miR-150/HIG2 axis
regulates the proliferation, invasion and the balance of iron
metabolism of hepatocellular carcinoma. Cell Physiol Biochem.
49:1403–1419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zheng J, Zhou Z, Qiu Y, Wang M, Yu H, Wu
Z, Wang X and Jiang X: A prognostic ferroptosis-related lncRNAs
signature associated with immune landscape and radiotherapy
response in glioma. Front Cell Dev Biol. 9:6755552021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C
and Xu S: LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress
during erastin-induced ferroptosis in HepG2 hepatocellular
carcinoma cells. Sci Rep. 9:161852019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Hu T, Zhang C, Tang Q, Su Y, Li B, Chen L,
Zhang Z, Cai T and Zhu Y: Variant G6PD levels promote tumor cell
proliferation or apoptosis via the STAT3/5 pathway in the human
melanoma xenograft mouse model. BMC Cancer. 13:2512013. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Su X, Yang Y, Yang Q, Pang B, Sun S, Wang
Y, Qiao Q, Guo C, Liu H and Pang Q: NOX4-derived ROS-induced
overexpression of FOXM1 regulates aerobic glycolysis in
glioblastoma. BMC Cancer. 21:11812021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Wang Z, Ding Y, Wang X, Lu S, Wang C, He
C, Wang L, Piao M, Chi G, Luo Y and Ge P: Pseudolaric acid B
triggers ferroptosis in glioma cells via activation of Nox4 and
inhibition of xCT. Cancer Lett. 428:21–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Chen B, Wang H, Lv C, Mao C and Cui Y:
Long non-coding RNA H19 protects against intracerebral hemorrhage
injuries via regulating microRNA-106b-5p/acyl-CoA synthetase long
chain family member 4 axis. Bioengineered. 12:4004–4015. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
He GN, Bao NR, Wang S, Xi M, Zhang TH and
Chen FS: Ketamine induces ferroptosis of liver cancer cells by
targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des Devel Ther.
15:3965–3978. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Yang Y, Tai W, Lu N, Li T, Liu Y, Wu W, Li
Z, Pu L, Zhao X, Zhang T and Dong Z: lncRNA ZFAS1 promotes lung
fibroblast-to-myofibroblast transition and ferroptosis via
functioning as a ceRNA through miR-150-5p/SLC38A1 axis. Aging
(Albany NY). 12:9085–9102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Chen L, Wang C, Sun H, Wang J, Liang Y,
Wang Y and Wong G: The bioinformatics toolbox for circRNA discovery
and analysis. Brief Bioinform. 22:1706–1728. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Liu Z, Wang Q, Wang X, Xu Z, Wei X and Li
J: Circular RNA cIARS regulates ferroptosis in HCC cells through
interacting with RNA binding protein ALKBH5. Cell Death Discov.
6:722020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhang HY, Zhang BW, Zhang ZB and Deng QJ:
Circular RNA TTBK2 regulates cell proliferation, invasion and
ferroptosis via miR-761/ITGB8 axis in glioma. Eur Rev Med Pharmacol
Sci. 24:2585–2600. 2020.PubMed/NCBI
|
|
150
|
Zhang S, Sun J, Gu M, Wang G and Wang X:
Circular RNA: A promising new star for the diagnosis and treatment
of colorectal cancer. Cancer Med. 10:8725–8740. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Qiao G, Zhang W and Dong K: Regulation of
ferroptosis by noncoding RNAs: A novel promise treatment in
esophageal squamous cell carcinoma. Mol Cell Biochem.
477:2193–2202. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Kinoshita C and Aoyama K: The role of
non-coding RNAs in the neuroprotective effects of glutathione. Int
J Mol Sci. 22:42452021. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zhang Y, Koppula P and Gan B: Regulation
of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell
Cycle. 18:773–783. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Wang X, Liu K, Gong H, Li D, Chu W, Zhao
D, Wang X and Xu D: Death by histone deacetylase inhibitor
quisinostat in tongue squamous cell carcinoma via apoptosis,
pyroptosis, and ferroptosis. Toxicol Appl Pharmacol.
410:1153632021. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
He C, Wang C, Liu H and Shan B: Kayadiol
exerted anticancer effects through p53-mediated ferroptosis in
NKTCL cells. BMC Cancer. 22:7242022. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Wang Y, Zhao Y, Wang H, Zhang C, Wang M,
Yang Y, Xu X and Hu Z: Histone demethylase KDM3B protects against
ferroptosis by upregulating SLC7A11. FEBS Open Bio. 10:637–643.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Liu J, Kang R and Tang D: Signaling
pathways and defense mechanisms of ferroptosis. FEBS J. Jun
6–2021.(Epub ahead of print). View Article : Google Scholar
|
|
158
|
Chen X, Kang R, Kroemer G and Tang D:
Broadening horizons: The role of ferroptosis in cancer. Nat Rev
Clin Oncol. 18:280–296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Kang R, Kroemer G and Tang D: The tumor
suppressor protein p53 and the ferroptosis network. Free Radic Biol
Med. 133:162–168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Galván I, Inácio Â, Dañino M, Corbí-Llopis
R, Monserrat MT and Bernabeu-Wittel J: High SLC7A11 expression in
normal skin of melanoma patients. Cancer Epidemiol. 62:1015822019.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Sui S, Zhang J, Xu S, Wang Q, Wang P and
Pang D: Ferritinophagy is required for the induction of ferroptosis
by the bromodomain protein BRD4 inhibitor (+)-JQ1 in cancer cells.
Cell Death Dis. 10:3312019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Liu T, Jiang L, Tavana O and Gu W: The
deubiquitylase OTUB1 mediates ferroptosis via stabilization of
SLC7A11. Cancer Res. 79:1913–1924. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Dai X, Lu L, Deng S, Meng J, Wan C, Huang
J, Sun Y, Hu Y, Wu B, Wu G, et al: USP7 targeting modulates
anti-tumor immune response by reprogramming tumor-associated
macrophages in lung cancer. Theranostics. 10:9332–9347. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Wang Y, Yang L, Zhang X, Cui W, Liu Y, Sun
QR, He Q, Zhao S, Zhang GA, Wang Y and Chen S: Epigenetic
regulation of ferroptosis by H2B monoubiquitination and p53. EMBO
Rep. 20:e475632019. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Lin PL, Tang HH, Wu SY, Shaw NS and Su CL:
Saponin formosanin C-induced ferritinophagy and ferroptosis in
human hepatocellular carcinoma cells. Antioxidants (Basel).
9:6822020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Xiao S, Liu X, Yuan L and Wang F: A
ferroptosis-related lncRNAs signature predicts prognosis and
therapeutic response of gastric cancer. Front Cell Dev Biol.
9:7366822021. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Wang Z, Zhang X, Tian X, Yang Y, Ma L,
Wang J and Yu Y: CREB stimulates GPX4 transcription to inhibit
ferroptosis in lung adenocarcinoma. Oncol Rep. 45:882021.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Zilka O, Poon JF and Pratt DA:
Radical-trapping antioxidant activity of copper and nickel
Bis(Thiosemicarbazone) complexes underlies their potency as
inhibitors of ferroptotic cell death. J Am Chem Soc.
143:19043–19057. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Hassannia B, Vandenabeele P and Vanden
Berghe T: Targeting ferroptosis to iron out cancer. Cancer Cell.
35:830–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Poon JF and Pratt DA: Recent insights on
hydrogen atom transfer in the inhibition of hydrocarbon
autoxidation. Acc Chem Res. 51:1996–2005. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Jiang L, Kon N, Li T, Wang SJ, Su T,
Hibshoosh H, Baer R and Gu W: Ferroptosis as a p53-mediated
activity during tumour suppression. Nature. 520:57–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Wang SJ, Li D, Ou Y, Jiang L, Chen Y, Zhao
Y and Gu W: Acetylation is crucial for p53-mediated ferroptosis and
tumor suppression. Cell Rep. 17:366–373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Nakamura T, Naguro I and Ichijo H: Iron
homeostasis and iron-regulated ROS in cell death, senescence and
human diseases. Biochim Biophys Acta Gen Subj. 1863:1398–1409.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Luo Y, Huang Q, He B, Liu Y, Huang S and
Xiao J: Regulation of ferroptosis by non-coding RNAs in the
development and treatment of cancer (Review). Oncol Rep. 45:29–48.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Pei Y, Qian Y, Wang H and Tan L:
Epigenetic regulation of ferroptosis-associated genes and its
implication in cancer therapy. Front Oncol. 12:7718702022.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Moloney JN and Cotter TG: ROS signalling
in the biology of cancer. Semin Cell Dev Biol. 80:50–64. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu
S and Tao Y: The epigenetic regulators and metabolic changes in
ferroptosis-associated cancer progression. Mol Cancer. 19:392020.
View Article : Google Scholar : PubMed/NCBI
|