Introduction
As a form of primary malignant bone tumor,
osteosarcoma (OS) is ranked first in the cause of cancer-associated
death in children and adolescents (1,2). OS is
derived from mesenchymal cells, characterized by a high recurrence
rate, early lung metastasis, and rapid infiltrating growth
(3). Surgical resection, adjuvant
chemotherapy, and small molecule targeted therapy have been used
for OS treatment (4,5); however, because of the high propensity
of metastasis and relapse the prognosis remains poor (6). Thus, it would be beneficial to
identify novel OS therapies by studying complex gene regulatory
networks. Circular RNAs (circRNAs) do not code for proteins and are
characterized by a covalently closed loop (7). CircRNAs are resistant to RNA
exonuclease digestion and therefore they are structurally stable
(8). Through sponging miRNAs,
circRNAs participate in the regulation of diverse cellular
processes, such as cell division, apoptosis, migration and
ferroptosis (9–12). CircRNAs exert vital functions in the
progression of various cancers including OS. For instance,
circRNA_103801 facilitates OS proliferation through regulating the
miR-338-3p/HIF-1/Rap1/PI3K-Akt signaling pathway (13), circRNA_0008035 silencing represses
OS metastasis by sponging miR-375 (14), and circMTO1 suppresses OS metastasis
by regulating the miR-630/KLF6 signaling (15). CircBLNK is a newly discovered
circRNA, located in chromosome 10, however, its functions in
tumorigenesis remain understudied.
MicroRNAs (miRNAs) are an abundant class of small
non-coding RNAs with 18–24 nucleotides in length, typically acting
as competing endogenous RNAs (ceRNAs) to suppress protein
translation or promote mRNA decay (16). Numerous studies have implicated
miRNAs in the regulation of tumorigenesis of malignancies. For
instance, miR-151a-3p regulates OS cell proliferation by targeting
RAB22A (17), miR-149 affects the
tumorigenesis of OS through targeting MSI2 (18) and miR-16b promotes OS cell
proliferation through regulating the PI3K/AKT signaling pathway
(19). MiR-188-3p acts as a ceRNA
in the tumorigenesis of various malignancies (20–22).
However, whether miR-188-3p could regulate the tumorigenesis of OS
remains unknown. Glutathione peroxidase 4 (GPX4) uses glutathione
as a cofactor to deplete membrane phospholipid hydroperoxides,
thereby maintaining cellular redox homeostasis (23). Abnormal activation of GPX4 can
induce ferroptosis (24), a
non-apoptotic form of cell death caused by iron-dependent
accumulation of toxic lipid peroxides (25). Furthermore, GPX4 has been implicated
in OS tumorigenesis (26,27). However, whether GPX4 could be
targeted by miRNA to regulate OS tumorigenesis remains
unstudied.
In the present study, the aim was to functionally
dissect the role of circBLNK in OS progression and to delineate the
relevant molecular mechanisms to gain new insights into the
etiology of OS.
Materials and methods
Clinical tissue samples
Tissues of OS and non-cancerous regions were
collected from 40 patients (age range, 16–54 years; mean age,
35±1.2 years; male/female ratio, 3:5) by surgery at the 970th
hospital of the PLA Joint Logistic Support Force between August
2017 and June 2021. No chemotherapy or radiotherapy was applied to
treat the patients before surgery. The tissue samples were verified
by two pathologists. All patients were informed of the research
design and written informed consent was provided by all patients.
The present study was approved (approval no. EC-H-2021-12-16) by
the Research Ethics Committee of the 970th hospital of the PLA
Joint Logistic Support Force (Yantai, China), which was in
accordance with the ethical standards formulated in the Helsinki
Declaration (28).
Cell culture and transfection
The 293T cell line was purchased from the American
Type Culture Collection, and four OS cell lines (HOS, SJSA-1, MG63
and U2OS) and the normal human osteoblast cell line (hFOB1.19) were
obtained from ScienCell Research Laboratories, Inc. Cells were
cultured in RPMI-1640 medium (HyClone; Cytiva) under a condition of
5% CO2 and 37°C. All media were supplemented with 10%
(v/v) fetal bovine serum (FBS; MilliporeSigma), 100 IU/ml
penicillin and 100 mg/ml streptomycin. Overexpression (OE)-vector,
small interfering (si)-NC, si-circBLNK, OE-GPX4, inhibitor control,
miR-188-3p inhibitor, mimic control, and miR-188-3p mimics were
provided by Shanghai GeneChem Co., Ltd. Transfection of cells with
a total of 20 nM of each indicated construct was implemented at
37°C for 15 min using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the
transfected cells were harvested to perform subsequent experiments.
The sequences used were as follows: si-NC:
5′-TTCTCCGAACGTGTCACGT-3′; si-circBLNK#1:
5′-AGATCTGTCTGAGAAGAAACA-3′; si-circBLNK#2:
5′-GATCTGTCTGAGAAGAAACAA-3′; miR-NC mimic:
5′-GUCGUAUCCAGUGCAGGGUCC-3′; miR-188-3p mimic:
5′-CUCCCACAUGCAGGGUUUGCA-3′; miR-NC inhibitor:
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-188-3p inhibitor:
5′-GAGGGUGUACGUCCCCAAACGU-3′.
RNase R treatment assay
Cytoplasmic and nuclear RNAs were separated using a
cytoplasmic and nuclear RNA separation kit according to the
manufacturer's protocol. Total RNAs were isolated by
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) from OS tissues and MG63 and U2OS cell lines according to the
manufacturer's protocol. For RNase R assay, RNase R (3 U/mg; cat.
no. ab286929; Abcam) was applied to the RNAs and incubated for 15
min at 37°C. Afterwards, the remaining RNAs were assessed by
reverse transcription-quantitative PCR (RT-qPCR).
RT-qPCR
Following RNA isolation by TRIzol®
reagent from MG63 and U2OS cells, cDNA was synthesized by a
High-Capacity cDNA Reverse Transcription kit (cat. no. 4368814;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instruction. The Thermal Cycler Dice System (Takara Bio, Inc.) with
the SYBR Green PCR Master Mix (Thermo Fisher Scientific, Inc.) were
used to perform qPCR. The reaction mixture was incubated at 95°C
for 2 min. The thermocycling program consisted of mainaining at
94°C for 2 min, followed by 30 cycles of 30 sec at 94°C, 30 sec at
56°C, and 60 sec at 72°C. The 2−ΔΔCq method was employed
to assess relative expression levels (29). The primers are listed in Table I.
 | Table I.The sequences of primers used for
reverse transcription-quantitative PCR. |
Table I.
The sequences of primers used for
reverse transcription-quantitative PCR.
| Gene name | Primer sequence
(5′→3′) |
|---|
| CircBLNK | F:
CAATGGTGGGGAATCAGTGT |
|
| R:
CTTCCATCTTCCCTCACAGC |
|
microRNA-188-3p | F:
CTCCCACATGCAGGGT |
|
| R:
GTCCAGTTTTTTTTTTTTTTTGCAA |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| GPX4 | F:
TCAGCAAGATCTGCGTGAAC |
|
| R:
TCAGCAAGATCTGCGTGAAC |
| GAPDH | F:
GTCAGTGGTGGACCTGACCT |
|
| R:
TGCTGTAGCCAAATTCGTTG |
Western blot analysis
The protein was extracted from MG63 and U2OS cells
as previously described (30).
Lysate samples (40 µg) were applied to a 10% SDS-PAGE gel, and
following electrophoresis, the separated protein was transferred to
a PVDF membrane (Invitrogen; Thermo Fisher Scientific, Inc.). After
treatment with 5% skim milk at 37°C for 2 h, the membrane was
treated with anti-GPX4 (1:1,000; cat. no. ab125066; Abcam) and
GAPDH (1:1,000; cat. no. 92310; Cell Signaling Technology, Inc.) at
4°C for 24 h. Subsequently, the membrane was treated with the IgG
H&L (HRP-conjugated; 1:10,000; cat. no. 7076S; Cell Signaling
Technology, Inc.) at 37°C for 2 h. The blot was finally developed
by ECL reagents (Amersham; Cytiva), and the band intensity was
measured using ImageJ version 4.1 (National Institutes of
Health).
Cell cytosolic/nuclear fractionation
assay
The nuclear and cytosolic fractions of MG63 and U2OS
cells were separated using the Cytoplasmic & Nuclear RNA
Purification Kit (100) (cat. no. NGB-37400; Norgen Biotek Corp.)
according to the manufacturer's protocol. The RNA was extracted
from the nuclear and cytosolic fractions. Afterwards, the
subcellular location of circBLNK in OS cells was inferred by its
abundance in different fractions.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was assessed by the CCK-8 assay.
In short, following the transfection of MG63 and U2OS cells with
corresponding constructs for 72 h, the cells were collected and
seeded into a 96-well plate (1×104 cells/well). After 0,
24, 48 and 72 h of cell culture, CCK-8 reagent (Thermo Fisher
Scientific, Inc.) (10 µl/well) was applied and incubated for 2 h at
37°C. The 450 nm absorbance was read by a microplate reader
(Bio-Rad Laboratories, Inc.).
MTT assay
The MTT assay was used to measure cell growth
capability in a 96-well plate. In short, 1×105 MG63 and
U2OS cells were seeded in each well. To assess the cell viability,
the cells were incubated for 24 h at 37°C. Subsequently, MTT
solution (20 µl/well, 5 mg/ml) (Thermo Fisher Scientific, Inc.) was
applied and incubated at 37°C for 4 h. Subsequently, dimethyl
sulfoxide (DMSO) (150 µl/well) (Invitrogen; Thermo Fisher
Scientific, Inc.) was added in order to dissolve the purple
formazan. Finally, the absorbance at 570 nm was calculated by an
ELISA browser (BioTek EL 800; Omega Bio-Tek, Inc.).
Clonogenic assay
The cells transfected with corresponding constructs
for 6 h were seeded into a six-well plate (1×104
cells/well). Then, MG63 and U2OS cells were cultured in complete
DMEM supplemented with 10% FBS at 37°C and 5% CO2. The
culture medium was replaced at 2-day intervals. After 14 days of
incubation, the colonies were washed twice with phosphate-buffered
saline (PBS) and fixed with 4% paraformaldehyde for 20 min. Next,
4% triformol (Invitrogen; Thermo Fisher Scientific, Inc.) was
applied to fix the cells followed by the staining with 0.1% crystal
violet (Beijing Solarbio Science & Technology Co.) for 20 min.
Finally, the colony number with >50 cells was counted under a
stereomicroscope (Olympus Corporation).
Flow cytometric analysis
Cell apoptosis of MG63 and U2OS cells was assessed
by the annexin V-FITC/PI Apoptosis Detection Reagent (cat. no.
88-8102-72; Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. A Beckman Coulter FACSCalibur (BD
Biosciences) flow cytometer coupled with the FlowJo 7.6.1 software
system (FlowJo LLC) was employed to detect apoptotic cells.
Bioinformatics analysis
The circRNA expression data was downloaded from Gene
Expression Omnibus (GEO; accession no. GSE140256; URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140256).
The binding sites between circBLNK and miR-183-3p, and the target
genes of miR-183-3p were predicted using CircInteractome (URL:
http://circinteractome.irp.nia.nih.gov/circular_rna.html),
MicroRNA Target Prediction Database (miRDB; http://mirdb.org/) and TargetScan 7.1 database
(www.targetscan.org).
Luciferase reporter assay
CircBLNK wild-type (circBLNK-WT), circBLNK mutant
(circBLNK-Mut), GPX4 wild-type (GPX4-WT), and
GPX4 mutant (GPX4-Mut) were subcloned into the
pGL3-basic vector (Shanghai GeneChem Co., Ltd.). Subsequently,
these constructs were co-transfected into respective 293T cell
lines with mimics of miR-183-3p or control using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After 72 h, the
transfected cells were harvested, and the luciferase activity of
the cells was calculated by the Luciferase Reporter Assay System
(Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity
was first normalized to that of Renilla luciferase from the
same sample.
RNA pull-down assay
The RNA pull-down assay was used to validate the
relationship between circBLNK and miR-188-3p. Briefly, the MG63 and
U2OS cells were transfected with circBLNK probe or NC probe for 48
h at 4°C. A total of ~1×107 cells were dissolved in the
soft lysis buffer plus 80 U/ml RNasin (Promega Corporation). The
probes were incubated at 4°C for 2 h with 50 µl C-1 magnetic beads
(Thermo Fisher Scientific, Inc.) to generate probe-coated beads and
then incubated at 4°C for 4 h with 0.7 ml RIPA lysis buffer (Thermo
Fisher Scientific, Inc.) to lyse MG63 and U2OS cells. After washing
in the lysis buffer (150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA and
0.5% NP40), RNA complexes were subjected to centrifugation at
11,100 × g for 10 min and then eluted by denaturation in 1X protein
loading buffer for 10 min at 100°C. Finally, miR-142-3p or
miR-142-5p transcripts were measured by RT-qPCR.
RNA immunoprecipitation (RIP)
assay
The RIP assay was used to assess the binding of
miR-188-3p to GPX4. After lysis in RIP the MG63 and U2OS cells
extract was incubated with magnetic beads conjugated with anti-Ago2
(1:2,000; cat. no. ab186733; Abcam) or normal anti-IgG (1:2,000;
cat. no. ab186733; Abcam) at 4°C for 24 h. Next, the magnetic beads
were treated with proteinase K, and the enrichment of miR-188-3p or
GPX4 in immunoprecipitated RNAs was determined with RT-qPCR.
Ferroptosis analysis
Ferroptosis analysis was used to measure the
ferroptosis cell death. In short, a mixture of 5 µM erastin
(Sigma-Aldrich; Merck KGaA), 1 µM ferrostatin-1 (cat. no.
347174-05-4; Sigma-Aldrich; Merck KGaA), 25 µM Z-VAD-FMK (cat. no.
V116; Sigma-Aldrich; Merck KGaA), and 20 µM necrosulfonamide (cat.
no. ab143839; Abcam) was used to treat MG63 and U2OS cells at 37°C
for 24 h. Afterwards, 0.1% DMSO (Invitrogen; Thermo Fisher
Scientific, Inc.) was applied to the cells followed by a 4 h
incubation at 37°C under serum-free conditions. Finally, the cell
viability was examined by the MTT assay.
Quantification of iron,
malondialdehyde (MDA) and lipid reactive oxygen species (ROS)
Levels of total iron and Fe2+ in MG63 and
U2OS cells were assessed by Iron Assay kit (cat. no. ab83366;
Abcam). Levels of MDA and lipid ROS were determined using Lipid
Peroxidation Assay Kit (cat. no. ab118970; Abcam) and Cellular ROS
Assay Kit (cat. no. ab186027; Abcam), respectively. The
aforementioned kits were used according to each manufacturer's
protocol.
Mitochondrial superoxide assay
MitoSOX™ Red (Thermo Fisher Scientific, Inc.) was
applied to measure the production of mitochondrial superoxide in
MG63 and U2OS cells. First, the working solution was prepared by
diluting 5 mM MitoSOX reagent stock solution (cat. no. M36005;
Thermo Fisher Scientific, Inc.) with Hanks' balanced salt solution
(HBSS; Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently,
the working solution was applied to cells and incubated in darkness
at 37°C for 10 min. The fluorescence intensity at FL2 was measured
by FACSCalibur (BD Biosciences).
Mitochondrial membrane potential
assay
A Mitochondrial Membrane Potential Kit (cat. no.
ab113850; Abcam) was used to measure the mitochondrial membrane
potential in MG63 and U2OS cells according to the manufacturer's
protocol.
Tumor formation experiment in nude
mice
Subcutaneous injection was performed by Vital River
Laboratories. MG63 cells (5×106) in 100 µl PBS were
injected into the breast fat pads of four-week-old (weight, 15–20
g) female BALB/c nude mice (n=12). The mice were raised under a
12/12-h light/dark cycle at 18–22°C and 50–60% humidity. Drinking
water was continuously provided and food was supplemented three
times a week. Both the research team and the veterinary staff
monitored animal health and behavior twice daily. Tumor size was
measured every three days from one week up to five weeks
post-injection. Tumor volume was calculated based on the formula
V=0.52 × ab2, where a represents tumor length
and b indicates tumor width. The humane endpoint about this
experiment was set to that the maximum size the tumors allowed to
grow in the mice before euthanasia was 2,000 mm3. At the
end of animal experiment, the maximum tumor weight/mice weight
observed was 7.4%. The maximum tumor diameter observed in this
study was 16.7 mm. The duration of the experiment was five weeks.
In strict accordance with the principles of animal welfare,
anesthesia was used to relieve pain during euthanasia under the
euthanasia by mask inhalation of isoflurane vaporized and
sacrificed by cervical dislocation. Isoflurane was used at 2% for
induction of anesthesia and at 1.5% for maintenance. The mice were
deemed dead when the following conditions occurred: Inability to
move, no spontaneous breathing, and no heartbeat. Each tumor weight
was documented. The animal protocol (approval no. EC-A-2022-5-23)
was approved by the Research Ethics Committee of the 970th hospital
of the PLA Joint Logistic Support Force (Yantai, China).
Statistical analysis
The data were analyzed and plotted as the mean ± SD
using the Prism 7.0 software (Dotmatics). Significance of
differences between two groups were compared with Student's t-test,
paired t-test was used in the statistical analysis of clinical
samples, and the other experiments adopted unpaired t-test to
analyze the difference. For multi-group data comparisons, one-way
ANOVA with Tukey's post hoc test was performed. The correlations
between the expression of circBLNK, miR-188-3p, and GPX4 in
OS tissues were analyzed using the Pearson's χ2 test.
The survival curves were established by the Kaplan-Meier plot (URL:
http://kmplot.com/analysis/).
P<0.05 was considered to indicate a statistically
significant difference.
Results
CircBLNK is upregulated in OS
tissues
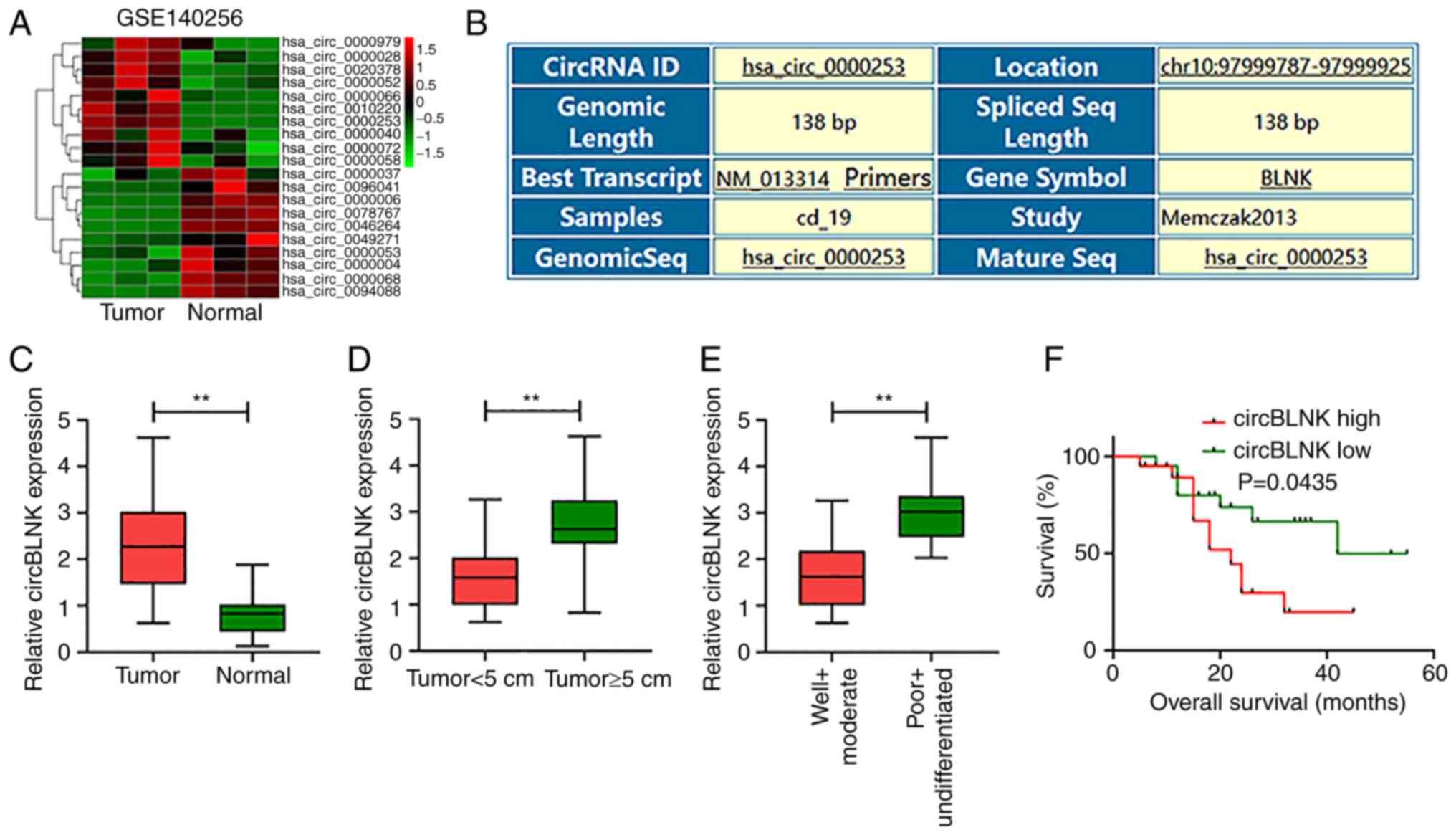
The circRNA expression data was downloaded from GEO
(accession no. GSE140256) (31) and
the differentially expressed circRNAs in OS were detected. CircBLNK
expression was observed significantly elevated in OS tissues (n=3)
relative to adjacent non-cancerous specimens (n=3) (Fig. 1A and B). Furthermore, circBLNK
expression in the 40 pairs of OS and adjacent non-cancerous tissues
was assessed by RT-qPCR. Consistently, circBLNK level in OS tissues
was significantly higher than in control tissues (Fig. 1C). Furthermore, a positive
association between circBLNK expression and tumor size (Fig. 1D) and cell differentiation (Fig. 1E) was detected. Based on the median
expression value of circBLNK, the 40 patients were divided into
circBLNK low and high expression groups. The Kaplan-Meier analysis
exhibited that high circBLNK expression was associated with poor
prognosis in OS patients (Fig. 1F).
Collectively, the results of the present study revealed the
anomalously high expression levels of circBLNK in OS tissues, which
are associated with poor OS prognosis.
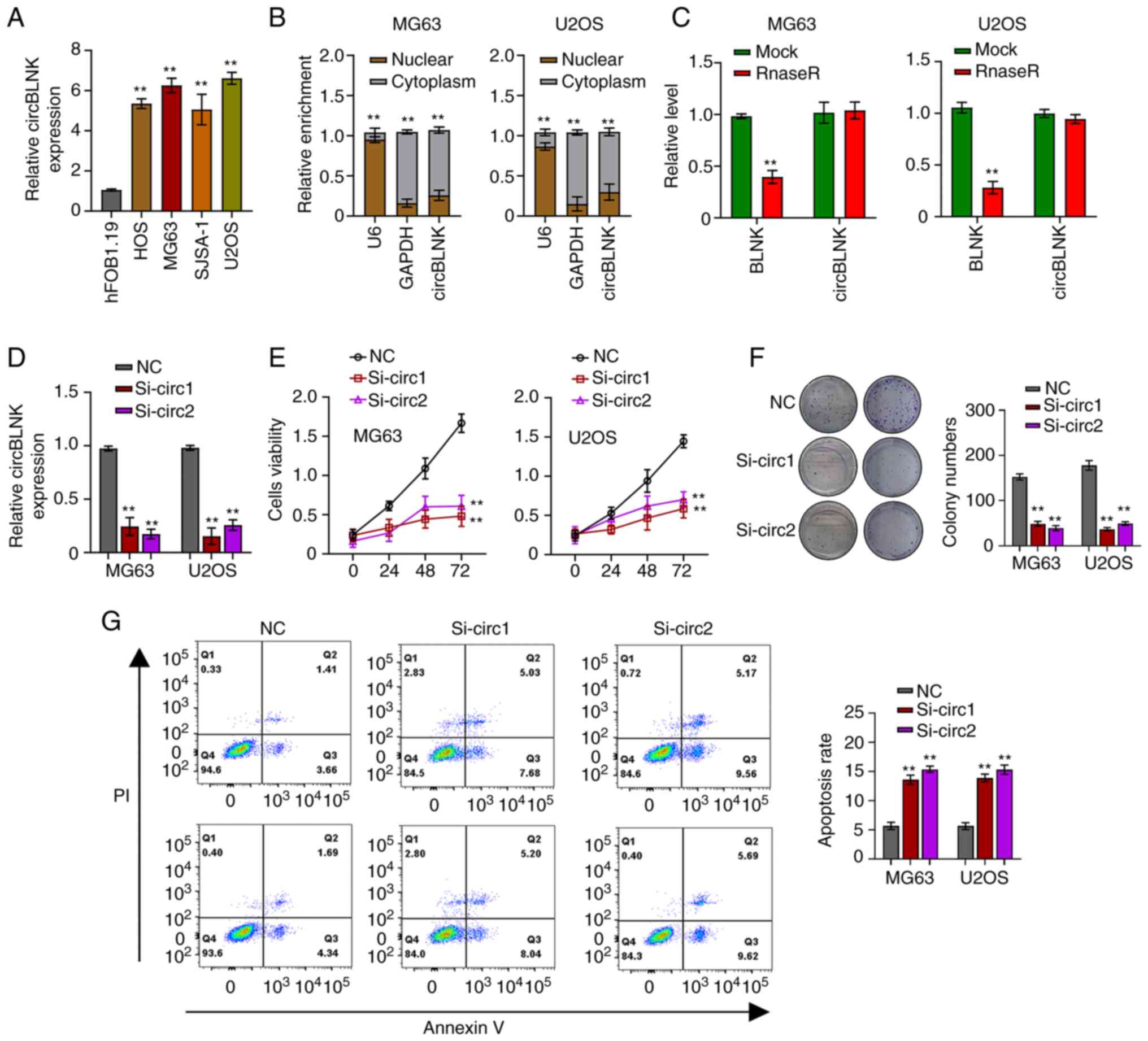
Upregulation of circBLNK in OS cells
promotes OS tumorigenesis
CircBLNK expression was further detected in four OS
cell lines (HOS, SJSA-1, MG63 and U2OS) and a human normal
osteoblast cell line (hFOB1.19). The four OS cell lines exhibited
significantly higher circBLNK levels than hFOB1.19, with MG63 and
U2OS cells exhibiting the highest (Fig.
2A). Afterwards, the subcellular localization of circBLNK in
MG63 and U2OS cells was detected using the cell cytosolic/nuclear
fractionation assay. The results revealed a specific cytoplasmic
localization of circBLNK (Fig. 2B).
Subsequently, the RNase R assay was used to evaluate the circular
nature of circBLNK and it was revealed that RNase R efficiently
digested liner BLNK but could not digest circBLNK (Fig. 2C). To dissect the role of circBLNK
in OS progression, circBLNK expression in MG63 and U2OS cells was
silenced by transfecting the cells with si-NC or si-circBLNK
(Fig. 2D). Next, the CCK-8 and
clonogenic assays were used to assess how circBLNK knockdown
affected the cell proliferation ability and it was detected that
the cell viability and colony-formation ability were suppressed
(Fig. 2E and F). On the other hand,
the cell apoptosis was enhanced when circBLNK expression was
silenced (Fig. 2G). Collectively,
the results of the present study revealed that circBLNK knockdown
suppresses cell proliferation and enhances cell apoptosis.
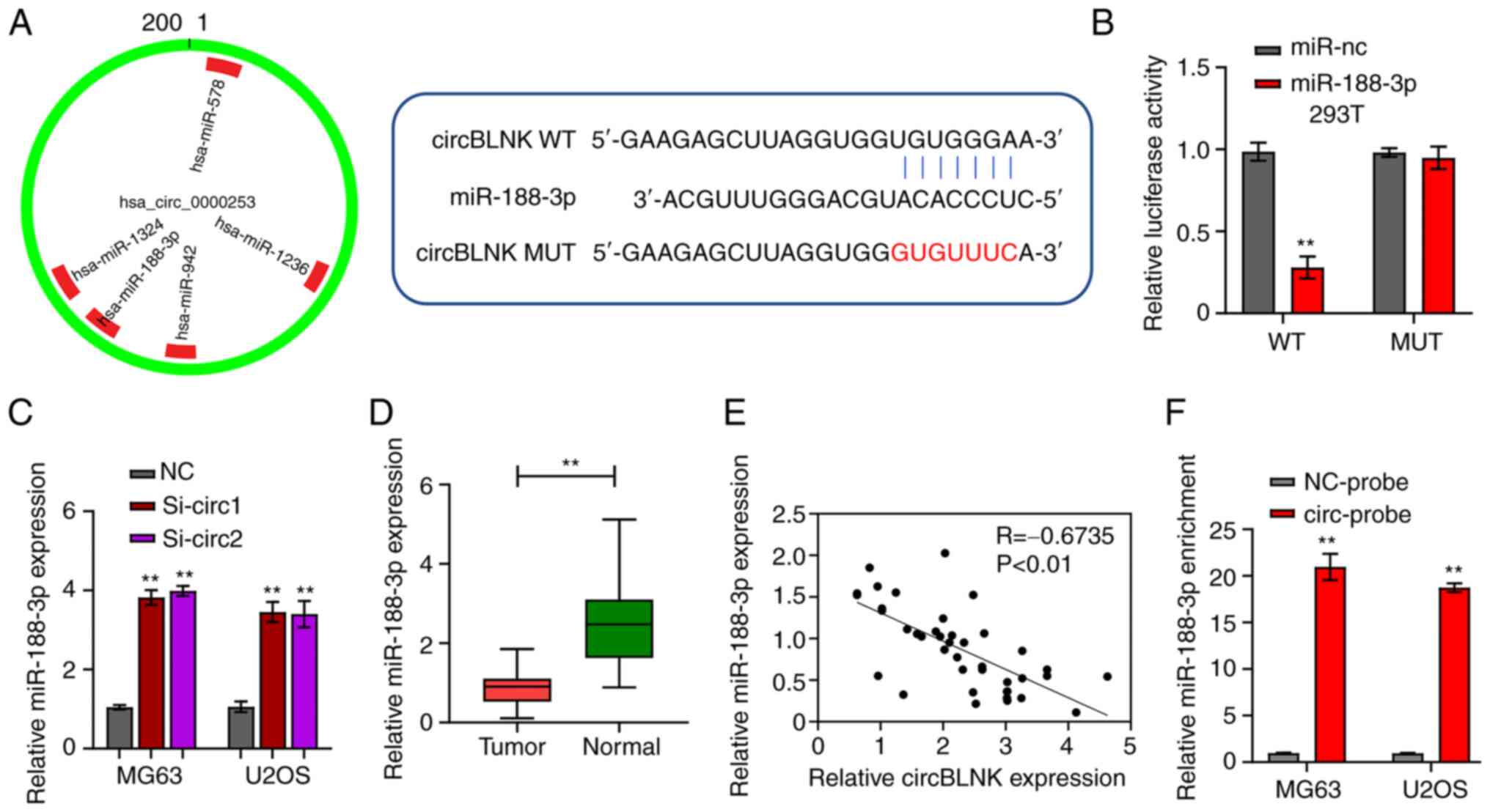
CircBLNK function as a sponge of
miR-188-3p
CircInteractome revealed that circBLNK could bind to
miR-188-3p (Fig. 3A). The
luciferase reporter and RNA pull-down assays were then performed to
check this possibility. Co-transfection of WT-circBLNK with
miR-188-3p mimics into 293T cells diminished the luciferase
reporter activity, whereas co-transfection of MUT-circBLNK with
miR-188-3p mimics retained the normal activity compared with the
co-transfection with mimic control (Fig. 3B). Furthermore, using RT-qPCR,
miR-188-3p expression in MG63 and U2OS cells transfected with si-NC
or si-circBLNK was determined. The results demonstrated that
knockdown of circBLNK increased miR-188-3p expression (Fig. 3C). In parallel, miR-188-3p
transcript abundance was determined in the 40 pairs of OS and
adjacent non-cancerous tissues. The abundance of miR-188-3p in OS
tissues was significantly lower than in adjacent non-cancerous
specimens (Fig. 3D). In addition,
circBLNK expression was detected to be negetively correlated with
miR-188-3p expression in OS tissues (Fig. 3E). These analyses suggested that
miR-188-3p expression is subjected to the regulation by circBLNK.
Next, for the collection of direct evidence supporting that
circBLNK targets miR-188-3p, the RNA pull-down assay was performed.
The assay detected the enrichment of miR-188-3p in the circBLNK
probe fraction compared with the NC probe fraction (Fig. 3F). Taken together, these findings
indicated that circBLNK regulates the OS tumorigenesis by sponging
miR-188-3p.
GPX4 is a direct target of
miR-188-3p
The miRDB revealed a high possibility that
miR-188-3p bind to GPX4 (Fig.
4A). The luciferase reporter assay was then performed to
validate this interaction. Co-transfection of miR-188-3p mimics and
WT-GPX4 resulted in weaker luciferase reporter activity in
293T cells, whereas the cells co-transfected with miR-188-3p mimics
and MUT-GPX4 showed normal luciferase reporter activity
(Fig. 4B). Moreover, the RIP assay
displayed that both miR-188-3p and GPX4 were effectively
pulled down by Ago2, indicating the direct interaction between them
(Fig. 4C and D). Afterwards,
GPX4 expression was measured in the 40 pairs of OS and
adjacent non-cancerous tissues by RT-qPCR. GPX4 expression was
significantly elevated in OS tissues (Fig. 4E). Additionally, a positive
correlation was detected between the abundance of circBLNK and
GPX4, whereas a negative correlation was found between the
abundance of miR-188-3p and GPX4 in OS tissues (Fig. 4F). Moreover, the levels of GPX4
transcripts and protein were all diminished in MG63 and U2OS cells
transfected with si-circBLNK, which was reversed with the presence
of miR-188-3p inhibitor (Fig. 4G and
H). Furthermore, circBLNK knockdown suppressed the
proliferation but enhanced the cell apoptosis of MG63 and U2OS
cells, while these effects were reversed with the presence of
miR-188-3p inhibitor (Fig. 4I and
J). In summary, the results of the present study demonstrated
that circBLNK promotes OS progression by regulating the
miR-188-3p/GPX4 pathway.
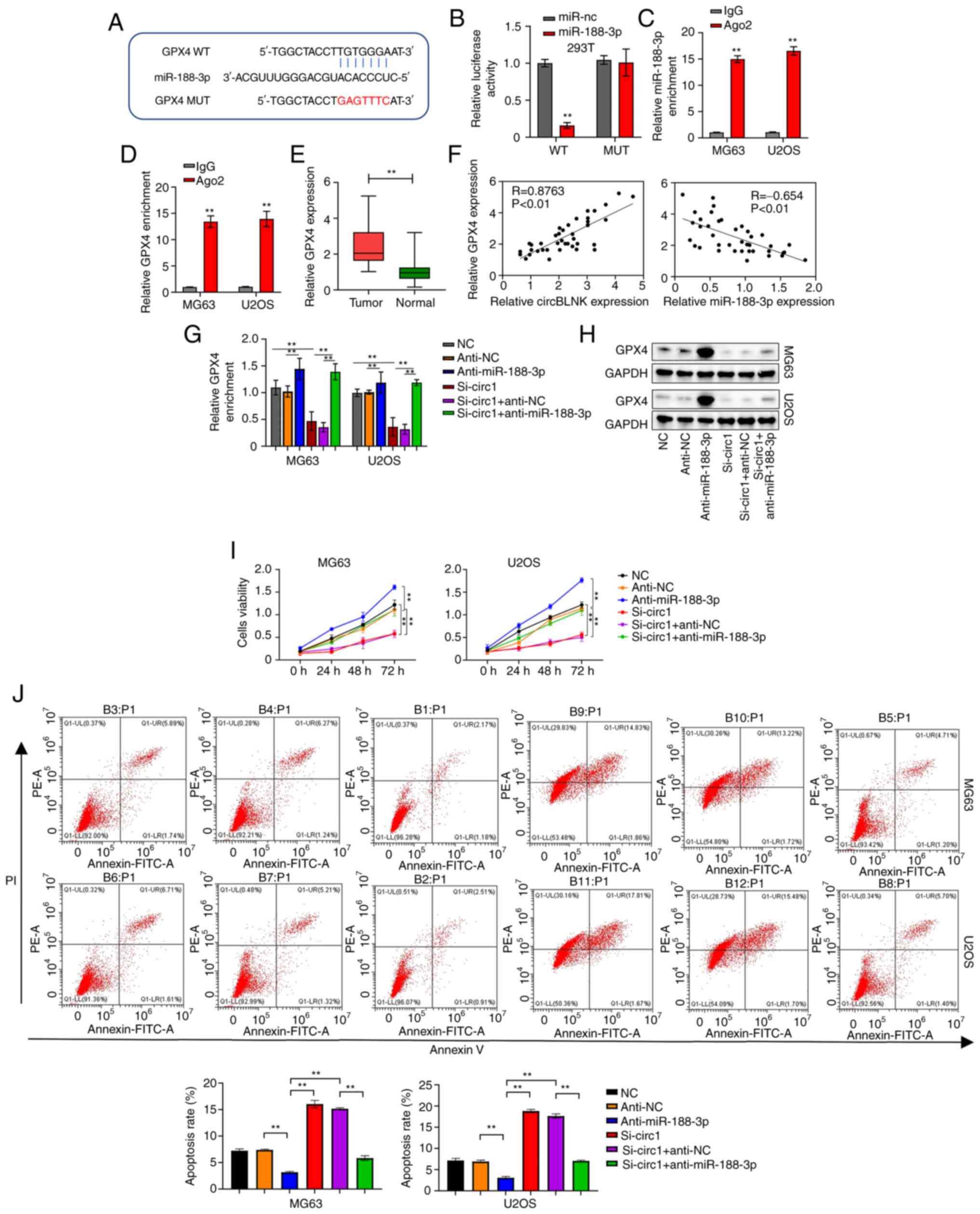 | Figure 4.GPX4 is a direct target of
miR-188-3p. (A) Complementary sequences of miR-188-3p and
GPX4 predicted by the miRDB database. (B) Relationship
between miR-188-3p and GPX4 confirmed by luciferase reporter assay.
(C) Interaction between miR-188-3p and GPX4 determined using RIP
assay. (D) Relationship between miR-188-3p and GPX4 assessed using
RIP assay. (E) GPX4 expression levels in 40 pairs of adjacent
non-cancerous and OS tissues was calculated using RT-qPCR. (F) The
correlation between the expression levels of circBLNK and GPX4 or
miR-188-3p and GPX4 in OS tissues was analyzed by the Pearson's
correlation coefficient. (G) GPX4 expression levels in OS cell
lines (MG63 and U2OS) transfected with indicated constructs (si-NC,
anti-NC, anti-miR-188-3p, si-circBLNK, si-circBLNK + anti-NC, and
si-circBLNK+anti-miR-188-3p) was calculated by RT-qPCR. (H) Protein
levels of GPX4 in OS cell lines (MG63 and U2OS) transfected with
indicated constructs (si-NC, anti-NC, anti-miR-188-3p, si-circBLNK,
si-circBLNK + inhibitor control, and si-circBLNK+miR-188-3p
inhibitor) calculated by western blot. (I) Cell proliferation in OS
cell lines (MG63 and U2OS) transfected with indicated constructs
(si-NC, anti-NC, anti-miR-188-3p, si-circBLNK, si-circBLNK +
anti-NC, and si-circBLNK + anti-miR-188-3p) was calculated by Cell
Counting Kit-8 assay. (J) Number of apoptotic cells in OS cell
lines (MG63 and U2OS) transfected with indicated constructs (si-NC,
anti-NC, anti-miR-188-3p, si-circBLNK, si-circBLNK + anti-NC and
si-circBLNK + anti-miR-188-3p) was calculated by flow cytometric
analysis. All experiments were carried out in triplicate.
**P<0.01. GPX4, glutathione peroxidase 4; miR, microRNA; miRDB,
MicroRNA Target Prediction Database; OS, osteosarcoma; RT-qPCR,
reverse transcription-quantitative PCR; RIP, RNA
immunoprecipitation; si-, small interfering; NC, negative
control. |
CircBLNK inhibits ferroptosis in OS
cells by regulating the miR-188-3p/GPX4 signaling
Finally, the regulation of ferroptosis by the
circBLNK/miR-188-3p/GPX4 signaling was investigated in OS cells.
First, with the use of MTT assay, erastin, a ferroptosis inducer,
was verified to activate cell death in MG63 and U2OS cells, whereas
ferrostain-1, a ferroptosis inhibitor, could counteract the effect
of erastin (Fig. 5A). Of note,
ZVAD-FMK (apoptosis inhibitor) and necrosulfonamide (necroptosis
inhibitor) did not affect the induction of cell death by erastin
(Fig. 5A). Afterwards, using the
MTT assay, the way circBLNK/miR-188-3p/GPX4 signaling affects
ferroptosis induction by erastin was investigated. The percentage
of cells undergoing cell death was revealed to be increased by
circBLNK knockdown, an effect that was reversed by
OE-GPX4-overexpressing cells (Fig.
5B). Since cell death is dependent on intracellular iron
(Fe2+) level, it was then assessed whether the
circBLNK/miR-188-3p/GPX4 signaling affects the intracellular
Fe2+ level in MG63 andU2OS cells. The concentration of
Fe2+ was demonstrated to be increased when circBLNK was
knocked down, which was rescued by GPX4-overexpressing cells
(Fig. 5C and D). Furthermore,
circBLNK knockdown resulted in an enhancement of MDA and lipid ROS
generation, which was reversed by GPX4-overexpressing cells
(Fig. 5E and F). In the cells with
circBLNK knockdown, the mitochondrial superoxide concentration was
increased, while the mitochondrial membrane potential was
diminished, which were eliminated by GPX4-overexpressing cells
(Fig. 5G and H). These results
suggested that circBLNK inhibits ferroptosis by regulating the
miR-188-3p/GPX4 signaling in OS cells.
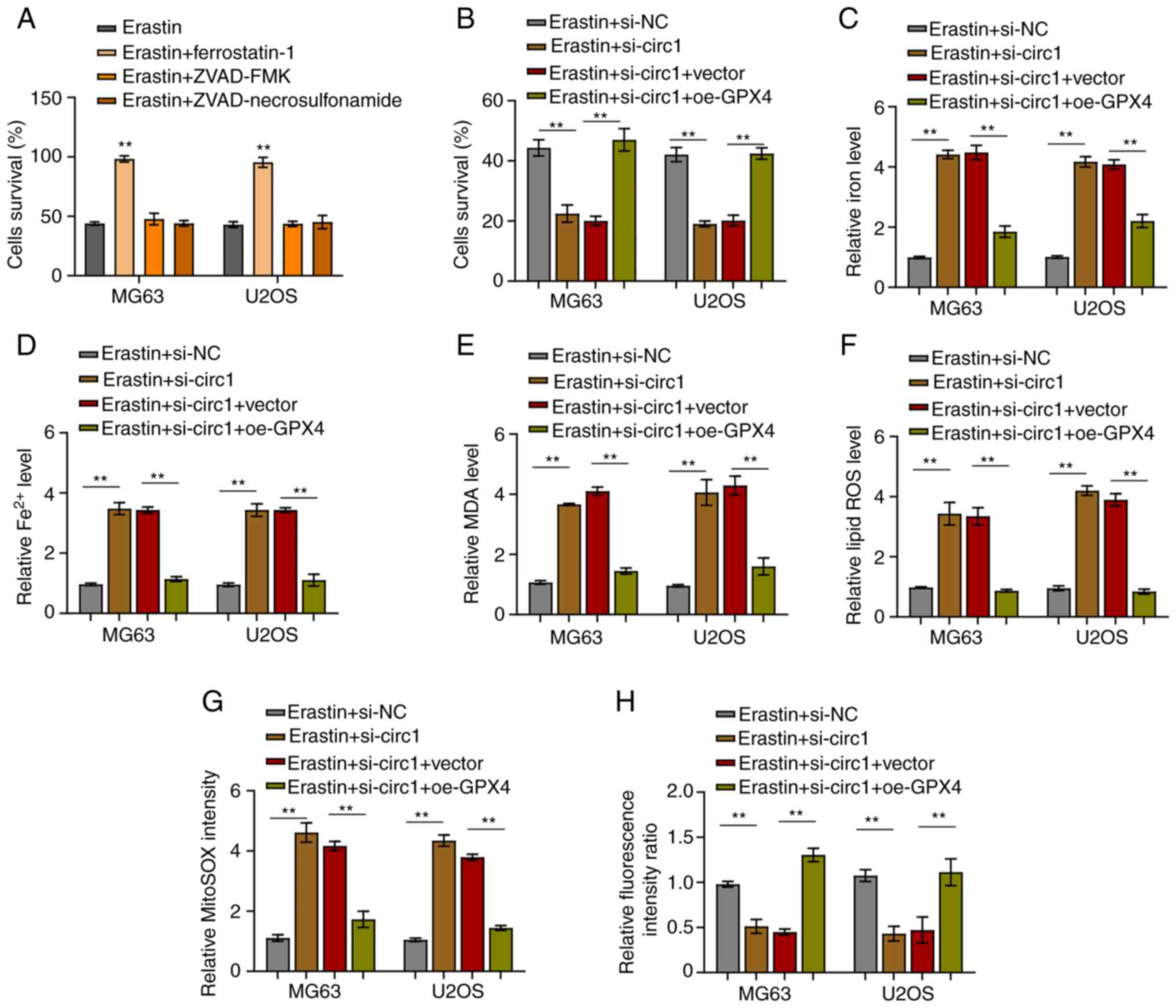 | Figure 5.CircBLNK inhibits ferroptosis in OS
cells through regulating the microRNA-188-3p/GPX4 signaling. (A)
Cell growth in OS cell lines (MG63 and U2OS) treated with indicated
constructs (Erastin, Erastin + ferrostatin-1, Erastin + ZVAD-FMK
and Erastin + necrosulfonamide) was evaluated by MTT assay. (B)
Cell growth in OS cell lines (MG63 and U2OS) treated with indicated
constructs (Erastin + si-NC, Erastin + si-circBLNK, Erastin +
si-circBLNK + OE-vector, and Erastin + si-circBLNK + OE-GPX4) was
assessed by MTT assay. (C) Expression level of total iron levels in
was determined through an iron assay kit. (D) Fe2+
accumulation was assessed by an iron assay kit. (E) MDA level was
determined by lipid peroxidation assay. (F) Lipid ROS level was
detected using flow cytometric analysis. (G) Mitochondrial
superoxide concentration was determined by mitochondrial superoxide
assay. (H) Mitochondrial membrane potential was evaluated using
mitochondrial membrane potential assay. All experiments were
carried out in triplicate. **P<0.01. Circ-, circular; OS,
osteosarcoma; si-, small interfering; NC, negative control; OE,
overexpression; MDA, malondialdehyde; ROS, reactive oxygen species;
GPX4, glutathione peroxidase 4. |
CircBLNK knockdown inhibits OS tumor
growth
To confirm circBLNK is a bona fide tumor
promoter in vivo, MG63 cells stably transfected with si-NC
or si-circBLNK were used to inject nude mice subcutaneously. Tumor
volume detection revealed a significant decrease in subcutaneous
tumor volume in the si-circBLNK group vs. the si-NC group (Fig. S1A). Similarly, circBLNK knockdown
significantly reduced the tumor weight (Fig. S1B). Collectively, the
aforementioned data suggested that circBLNK promotes OS tumor
growth in vivo.
Discussion
With the advent of high-throughput genomic and RNA
sequencing, a large array of circRNAs have been identified. Over
the past decades, the pivotal role of circRNAs in malignant tumor
tumorigenesis has been well documented (32,33).
In the present study, the role of circBLNK in OS progression was
molecularly dissected. CircBLNK was elevated in both OS tissues and
cell lines, facilitating the proliferation while impairing the
apoptosis and ferroptosis of OS cells. Altogether, the
circBLNK/miR-188-3p/GPX4 axis was identified as a new
transcriptional regulatory network for OS progression.
CircBLNK is a newly discovered circRNA, located in
chromosome 10. In the present study, circBLNK expression was
revealed to be elevated in OS tissues and this was associated with
large tumor size, low cell differentiation and poor prognosis.
Then, the use of RT-qPCR confirmed the elevated circBLNK expression
in both OS tissues and cell lines. Moreover, it was observed that
circBLNK knockdown impaired OS cell proliferation while enhancing
cell apoptosis. These results suggested that circBLNK may play a
facilitating role in OS tumorigenesis and progression.
CircRNAs exert their functions through sponging
miRNAs (34,35). It was observed that circRNAs are
involved in OS progression by sponging miRNAs. For instance,
circCAMSAP1 promotes OS development through sponging miR-145-5p
(36), hsa_circ_0008259 regulates
OS progression by sponging miR-21-5p (37), circEIF4G2 accelerates OS
tumorigenesis and progression by sponging miR-218 (38). In the present study, it was
demonstrated that miR-188-3p is the target for circBLNK. MiR-188-3p
expression was downregulated in many malignant tumor tissues
(39–41). Congruent with the aforementioned
findings of previous studies, it was also observed that miR-188-3p
was downregulated in OS tissues. In addition, the results of the
present study confirmed that circBLNK deficiency impaired OS cell
proliferation, while enhancing cell apoptosis, and both phenotypes
could be reversed by inhibiting miR-188-3p. Collectively, these
results demonstrated that circBLNK regulates the OS tumorigenesis
through sponging miR-188-3p.
CircRNAs typically act as ceRNAs to modulate the
expression of target miRNA (42).
In the present study, through a combination of bioinformatics
prediction, luciferase reporter assay, and RIP assay, GPX4
was demonstrated as the target gene of miR-188-3p. Furthermore, the
level of GPX4 transcripts was determined to be elevated in
OS tissues. Moreover, a positive correlation of GPX4
expression with circBLNK expression was observed, and a negative
correlation between GPX4 expression and miR-188-3p
expression was also identified. Thus, it was demonstrated that
circBLNK functions as a ceRNA for miR-188-3p to regulate GPX4
expression.
Ferroptosis has emerged as a new form of cell death
mediated by peroxidation of ROS and lipid (43). Ferroptosis-based therapy has been
proposed as an alternative treatment for malignant tumor treatment
(44). Sorafenib, an FDA-approved
small molecule drug for cancer therapy, is an inducer of
ferroptosis (45). Other than
Sorafenib, gene therapies based on ferroptosis-associated
nanomaterials are undergoing development (46). It has been revealed that ferroptosis
is tightly associated with the progression of diverse malignant
tumors including OS (27,47,48).
In the present study, it was identified that circBLNK silencing
augmented ferroptosis as reflected by the elevated levels of
intracellular Fe2+, MDA, lipid ROS and mitochondrial
superoxide as well as the diminished mitochondrial membrane
potential. This enhancement of ferroptosis by circBLNK silencing
could be eliminated in OE-GPX4-overexpressing cells. Collectively,
these results demonstrated that circBLNK inhibits OS cell
ferroptosis through the miR-188-3p/GPX4 signaling. In conclusion,
the present study investigated the upregulation of circBLNK in OS
tissues and cells, which predicts poor patient prognosis.
Mechanistically, circBLNK functions as a ceRNA to arrest miR-188-3p
expression and to upregulate GPX4 expression, thereby ultimately
promoting OS tumorigenesis. Meanwhile, the present study provided a
potential molecular target for OS early diagnosis and therapeutic
interventions.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by PLA Youth Training Project
for Medical Science (grant no. 17QNP015).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DXH and WPS made substantial contributions to
conception and design. ZJL, YL and CBW performed the experiments
and acquired the data. ZJL and CBW analyzed the data. DXH and YL
drafted the manuscript. ZJL and CBW critically revised the study
for important intellectual content. DXH and WPS confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript and agreed to be accountable for
all aspects of the work.
Ethics approval and consent to
participate
Human studies (approval no. EC-H-2021-12-16) and
animal experiments (approval no. EC-A-2022-5-23) were approved by
the Research Ethics Committee of the 970th hospital of the PLA
Joint Logistic Support Force (Yantai, China). All patients were
informed of the research design and written informed consent was
provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Noone AM, Cronin KA, Altekruse SF,
Howlader N, Lewis DR, Petkov VI and Penberthy L: Cancer incidence
and survival trends by subtype using data from the surveillance
epidemiology and end results program, 1992–2013. Cancer Epidemiol
Biomarkers Prev. 26:632–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rainusso N, Wang LL and Yustein JT: The
adolescent and young adult with cancer: State of the Art-bone
tumors. Curr Oncol Rep. 15:296–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bishop MW, Janeway KA and Gorlick R:
Future directions in the treatment of osteosarcoma. Curr Opin
Pediatr. 28:26–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Otoukesh B, Boddouhi B, Moghtadaei M,
Kaghazian P and Kaghazian M: Novel molecular insights and new
therapeutic strategies in osteosarcoma. Cancer Cell Int.
18:1582018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Sun D, Pu W, Wang J and Peng Y:
Circular RNAs in cancer: Biogenesis, function, and clinical
significance. Trends Cancer. 6:319–336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Z, Liu F, Wang F, Yang X and Guo W:
CircZNF609 promotes cell proliferation, migration, invasion, and
glycolysis in nasopharyngeal carcinoma through regulating HRAS via
miR-338-3p. Mol Cell Biochem. 476:175–186. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan C, Qu H, Xiong F, Tang Y, Tang T,
Zhang L, Mo Y, Li X, Guo C, Zhang S, et al: CircARHGAP12 promotes
nasopharyngeal carcinoma migration and invasion via ezrin-mediated
cytoskeletal remodeling. Cancer Lett. 496:41–56. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang L, Tong X, Zhou Z, Wang S, Lei Z,
Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al: Circular RNA
hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced
epithelial-mesenchymal transition and metastasis by controlling
TIF1γ in non-small cell lung cancer. Mol Cancer. 17:1402018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li C, Tian Y, Liang Y and Li Q: Retraction
note to: Circ_0008035 contributes to cell proliferation and
inhibits apoptosis and ferroptosis in gastric cancer via
miR-599/EIF4A1 axis. Cancer Cell Int. 21:4162021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li ZQ, Wang Z, Zhang Y, Lu C, Ding QL, Ren
R, Cheng BB and Lou LX: CircRNA_103801 accelerates proliferation of
osteosarcoma cells by sponging miR-338-3p and regulating
HIF-1/Rap1/PI3K-Akt pathway. J Biol Regul Homeost Agents.
35:1021–1028. 2021.PubMed/NCBI
|
|
14
|
Gong G, Han Z, Wang W, Xu Q and Zhang J:
Silencing hsa_circRNA_0008035 exerted repressive function on
osteosarcoma cell growth and migration by upregulating
microRNA-375. Cell Cycle. 19:2139–2147. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu DY, Li Z, Zhang K, Jiao N, Lu DG, Zhou
DW, Meng YB and Sun L: Circular RNA CircMTO1 suppressed
proliferation and metastasis of osteosarcoma through miR-630/KLF6
axis. Eur Rev Med Pharmacol Sci. 25:86–93. 2021.PubMed/NCBI
|
|
16
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng S, Jiang F, Ge D, Tang J, Chen H,
Yang J, Yao Y, Yan J, Qiu J, Yin Z, et al: LncRNA
SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of
osteosarcoma. Biomed Pharmacother. 112:1086952019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang W, Li JZ, Tai QY, Tang JJ, Huang YH
and Gao SB: LncRNA DANCR regulates osteosarcoma migration and
invasion by targeting miR-149/MSI2 axis. Eur Rev Med Pharmacol Sci.
24:6551–6560. 2020.PubMed/NCBI
|
|
19
|
Xu M, Zhang YY, Wang HF and Yang GS: The
expression and function of miRNA-106 in pediatric osteosarcoma. Eur
Rev Med Pharmacol Sci. 21:715–722. 2017.PubMed/NCBI
|
|
20
|
Luo Z, Fan Y, Liu X, Liu S, Kong X, Ding
Z, Li Y and Wei L: MiR-188-3p and miR-133b suppress cell
proliferation in human hepatocellular carcinoma via
post-transcriptional suppression of NDRG1. Technol Cancer Res
Treat. 20:153303382110330742021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pei J, Zhang S, Yang X, Han C, Pan Y, Li
J, Wang Z, Sun C and Zhang J: Long non-coding RNA RP11-283G6.5
confines breast cancer development through modulating
miR-188-3p/TMED3/Wnt/β-catenin signalling. RNA Biol. 18 (Suppl
1):S287–S302. 2021. View Article : Google Scholar
|
|
22
|
Pei J, Zhang J, Yang X, Wu Z, Sun C, Wang
Z and Wang B: TMED3 promotes cell proliferation and motility in
breast cancer and is negatively modulated by miR-188-3p. Cancer
Cell Int. 19:752019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seiler A, Schneider M, Förster H, Roth S,
Wirth EK, Culmsee C, Plesnila N, Kremmer E, Rådmark O, Wurst W, et
al: Glutathione peroxidase 4 senses and translates oxidative stress
into 12/15-lipoxygenase dependent- and AIF-mediated cell death.
Cell Metab. 8:237–248. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Z, Chen L, Wang C, Zhang L and Xu W:
MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through
inhibiting GPX4. Free Radic Res. 55:1119–1129. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Q and Wang K: The induction of
ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the
sensitivity of osteosarcoma cells to cisplatin. Cell Biol Int.
43:1245–1256. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Issue Information-declaration of Helsinki.
J Bone Miner Res. 34:BM i–BM ii. 2019.
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng X, Huang M, Xing L, Yang R, Wang X,
Jiang R, Zhang L and Chen J: The circRNA circSEPT9 mediated by E2F1
and EIF4A3 facilitates the carcinogenesis and development of
triple-negative breast cancer. Mol Cancer. 19:732020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu L, Duan J, Li M, Zhou C and Wang Q:
Circ_0000253 promotes the progression of osteosarcoma via the
miR-1236-3p/SP1 axis. J Pharm Pharmacol. 75:227–235. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang D, Yang S, Wang H, Wang J, Zhang Q,
Zhou S, He Y, Zhang H, Deng F, Xu H, et al: The progress of
circular RNAs in various tumors. Am J Transl Res. 10:1571–1582.
2018.PubMed/NCBI
|
|
34
|
Yang Y, Yujiao W, Fang W, Linhui Y, Ziqi
G, Zhichen W, Zirui W and Shengwang W: The roles of miRNA, lncRNA
and circRNA in the development of osteoporosis. Biological Res.
53:402020. View Article : Google Scholar
|
|
35
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Z, Xu W, Zhang D, Chu J, Shen S, Ma
Y, Wang Q, Liu G, Yao T, Huang Y, et al: circCAMSAP1 promotes
osteosarcoma progression and metastasis by sponging miR-145-5p and
regulating FLI1 expression. Mol Ther Nucleic Acids. 23:1120–1135.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guan K, Liu S, Duan K, Zhang X, Liu H, Xu
B, Wang X and Jin X: Hsa_circ_0008259 modulates miR-21-5p and PDCD4
expression to restrain osteosarcoma progression. Aging (Albany NY).
13:25484–25495. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin E, Liu S, Xiang W, Zhang H and Xie C:
CircEIF4G2 Promotes Tumorigenesis and Progression of Osteosarcoma
by Sponging miR-218. Biomed Res Int. 2020:83869362020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Meng L, Jiang YP, Zhu J and Li B:
MiR-188-3p/GPR26 modulation functions as a potential regulator in
manipulating glioma cell properties. Neurol Res. 42:222–227. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pichler M, Stiegelbauer V,
Vychytilova-Faltejskova P, Ivan C, Ling H, Winter E, Zhang X,
Goblirsch M, Wulf-Goldenberg A, Ohtsuka M, et al: Genome-Wide miRNA
analysis identifies miR-188-3p as a novel prognostic marker and
molecular factor involved in colorectal carcinogenesis. Clin Cancer
Res. 23:1323–1333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao F, Han J, Wang Y, Jia L, Luo W and
Zeng Y: Circ_0109291 promotes cisplatin resistance of oral squamous
cell carcinoma by sponging miR-188-3p to increase ABCB1 expression.
Cancer Biother Radiopharm. 37:233–245. 2022.PubMed/NCBI
|
|
42
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hassannia B, Vandenabeele P and Vanden
Berghe T: Targeting ferroptosis to iron out cancer. Cancer Cell.
35:830–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen X, Kang R, Kroemer G and Tang D:
Broadening horizons: The role of ferroptosis in cancer. Nat Rev
Clin Oncol. 18:280–296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liang C, Zhang X, Yang M and Dong X:
Recent progress in ferroptosis inducers for cancer therapy. Adv
Mater. 31:e19041972019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R
and Tang D: Activation of the p62-Keap1-NRF2 pathway protects
against ferroptosis in hepatocellular carcinoma cells. Hepatology.
63:173–184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lei T, Qian H, Lei P and Hu Y:
Ferroptosis-related gene signature associates with immunity and
predicts prognosis accurately in patients with osteosarcoma. Cancer
Sci. 112:4785–4798. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Luo Y, Gao X, Zou L, Lei M, Feng J and Hu
Z: Bavachin induces ferroptosis through the STAT3/P53/SLC7A11 axis
in osteosarcoma cells. Oxid Med Cell Longev. 2021:17834852021.
View Article : Google Scholar : PubMed/NCBI
|