Introduction
Conventional monoclonal antibodies, mostly IgG1s,
have been widely used as therapies for diseases that are difficult
to treat (1,2); however, they have not been effective
for cancer treatment. Several strategies for improving the function
of therapeutic antibodies have been explored, such as designing
non-natural antibody formats, including fusion antibodies,
multivalent antibodies and multispecific antibodies, as well as
their combinations (3,4). Bispecific antibodies (bsAbs) among
multispecific antibodies are a practical non-natural antibody
format and have been approved by the United States Food and Drug
Administration (5). However, only
four cancer therapeutic bsAbs have been approved by the Food and
Drug Administration; thus, technologies are needed to accelerate
the development of highly functional non-natural antibodies for
treating cancer.
Multispecific antibodies were recently developed for
treating viral infections and cancer (6). For example, a trispecific antibody
targeting different epitopes on the envelope of human
immunodeficiency virus-1 showed antigen-binding ability and reduced
the effects of viral mutations that typically make therapy
challenging (7). Studies on cancer
therapeutic multispecific antibodies have recently increased
(8). For example, a previous study
revealed that trispecific antibodies targeting different targets in
cancer cells inhibit cancer growth (9). Multispecific antibodies designed to
redirect some types of immune cells as effector cells toward cancer
cells have been widely studied especially for developing bsAbs. As
effector cells, T cells and NK cells are often used to construct
multispecific antibodies (10,11),
due to their high abundance in the blood and strong antitumor
effects.
In previous studies by the authors, the development
and efficacy of bsAbs that recruit T cells or NK cells: Ex3
targeting epidermal growth factor receptor (EGFR) on tumor cells
and CD3 on T cells (12,13) and Ex16 targeting EGFR and CD16 on NK
cells, were reported (14,15). T cells and NK cells also exist in
solid tumors as tumor-infiltrating lymphocytes (TILs); however, a
major challenge in cancer therapy is exhaustion of these effector
cells (16). To activate such
exhausted immune cells, co-stimulatory molecules or agonist
antibodies with co-stimulatory signals have been mounted on bsAbs
that recruit immune cells to design multispecific antibodies with
strong therapeutic effects (17,18).
Another concern is the differences in the abundance ratios of T
cells and NK cells as TILs, depending on the individual cancer,
stage and malignancy (19).
Combination therapy with bsAbs that recruit T cells and NK cells
may induce therapeutic effects independent of the TIL populations;
however, administration of two biologicals is cost-ineffective due
to the high production costs of conventional therapeutic
antibodies. Therefore, the present study focused on the development
of novel trispecific antibodies recruiting both T and NK cells.
In the present study, three Fc-fused trispecific
antibodies were designed, namely T-cell and NK-cell engagers
(TaKEs), which target EGFR, CD3 and CD16. Among them, TaKE1-Fc was
circularly connected with four variable fragments (Fvs) from three
different antibody clones. Using highly purified TaKE1 after
removing the Fc region, its trispecificity via its ability to bind
cancer cells, T cells and NK cells, as well as its comparable or
stronger effects than those of the combination of the two bsAbs
were confirmed. Functional trispecific antibodies recruiting both T
cells and NK cells were created and a novel format for the
development of trispecific antibodies was provided.
Materials and methods
Construction of expression vectors and
preparation of three types of TaKE-Fc
It has been previously reported that bsAbs use the
variable heavy (VH) and variable light (VL) regions of the
humanized anti-CD3 antibody OKT3 (12), humanized anti-EGFR antibody 528
(12), mouse anti-CD16 antibody 3G8
(14) and mouse anti-EGFR antibody
225 (20). In the present study,
three trispecific antibodies (TaKEs) were designed (Fig. 1). Corresponding gene expression
cassettes were designed based on the amino acid sequences of the VH
and VL regions and human IgG1 Fc, synthesized, and then inserted
into the pCAGGS expression vector (21). All constructed gene sequences for
TaKE-Fcs are indicated in Fig. S1,
Fig. S2, Fig. S3. All TaKE-Fcs were expressed
transiently using Expi293 Expression System (Thermo Fisher
Scientific, Inc.) and purified using protein A chromatography
(rProtein A Sepharose™ Fast Flow; Cytiva) according to the
manufacturer's protocol. Briefly, the column was equilibrated with
Tris-HCl buffer containing 200 mM NaCl (pH 8.0). After the culture
supernatant was loaded onto the column, the column was washed with
Tris-HCl buffer containing 200 mM NaCl (pH 8.0). TaKE-Fcs were
eluted using Pierce IgG Elution Buffer (Thermo Fisher Scientific,
Inc.).
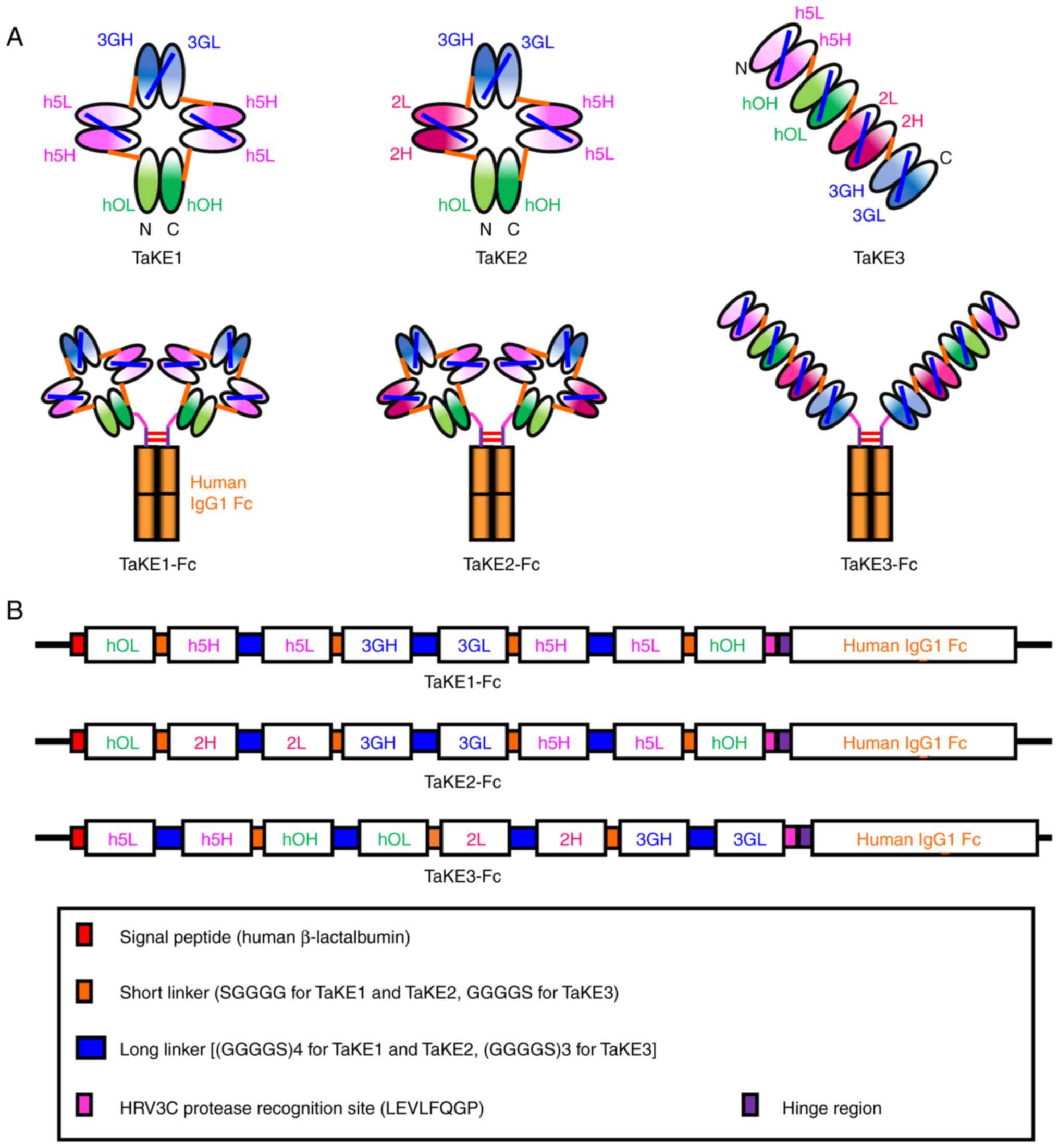 | Figure 1.Construction of TaKEs. (A) Schematic
diagrams of three types of TaKEs and their human IgG1 Fc fusion
formats. (B) Schematic diagrams of the expression vectors for TaKEs
with Fc. TaKE, T cell and natural killer (NK) cell engager; VH,
variable heavy; VL, variable light; hOH, the VH regions of the
humanized anti-CD3 antibody OKT3; hOL, the VL regions of the
humanized anti-CD3 antibody OKT3; h5H, the VH regions of the
humanized anti-EGFR antibody 528; h5L, the VL regions of the
humanized anti-EGFR antibody 528; 3GH, the VH regions of the mouse
anti-CD16 antibody 3G8; 3GL, the VL regions of the mouse anti-CD16
antibody 3G8; 2H, the VH regions of the mouse anti-EGFR antibody
225; 2L, the VL regions of the mouse anti-EGFR antibody 225. |
Preparation of TaKE1 and bsAbs
To prepare each TaKE1 without Fc, protein A
chromatography-purified TaKE1-Fc was digested with HRV3C protease
fused to glutathione S-transferase (PreScission protease;
Cytiva), according to the manufacturer's protocol and a previous
study by the authors (22). The
protease was removed on a Glutathione Sepharose 4 B column
(Cytiva), and the flow-through was loaded onto the protein A column
again to remove the digested Fc and undigested Fc fusion protein.
Gel filtration was performed using a Superdex 200 Increase column
(10/300; Cytiva) to fractionate the TaKE1 monomers. Purity and
successful preparation were confirmed using sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; SuperSep™ Ace
10–20%, 17 well; FUJIFILM Wako Pure Chemical Corporation) under
reducing conditions. Ex3-scDb-LH, a bispecific single-chain diabody
targeting EGFR and CD3 (22), was
prepared from its Fc fusion format in a similar manner.
Ex16-scDb-HL, a bispecific single-chain diabody targeting EGFR and
CD16, was prepared using the B. choshinensis expression
system (ProteinExpress Co., Ltd.), as previously reported (15). After purification of Ex16-scDb-HL
using immobilized metal affinity chromatography directly from the
bacterial supernatant, gel filtration was performed using a
Superdex 200 Increase column to fractionate the monomer.
Cell lines
Human bile duct carcinoma (TFK-1) was established by
the authors (23). CD3-positive
lymphokine-activated killer cells with the T-cell phenotype (T-LAK)
cells were induced from ethically collected peripheral blood
mononuclear cells (CTL-UP1; Cellular Technology Limited) as
previously described (24). These
cell lines were cultured in a humidified environment (37°C, 5%
CO2) in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA)
supplemented with 10% fetal bovine serum (Biowest), 100 U/ml
penicillin, and 100 µg/ml streptomycin. The NK92/CD16A cell line
was kindly provided by Professor Tachibana of Osaka Metropolitan
University (Osaka, Japan) and cultured in a humidified environment
(37°C, 5% CO2) in MyeloCult H5100 medium (StemCell
Technologies, Inc.) supplemented with 100 U/ml of recombinant human
interleukin-2 (IMUNACE 35 for Injection; Shionogi & Co., Ltd.)
(25).
Flow cytometric analysis
For binding analysis of TaKE1, TFK-1 cells, T-LAK
cells and NK92/CD16A cells were used as EGFR-positive,
CD3-positive, and CD16-positive cell lines, respectively. Each cell
line was incubated with TaKE1 (40 pmol) for 30 min on ice. After
being washed with PBS, rabbit anti-Ex3 serum, provided by
Immuno-Biological Laboratories Co., Ltd. through polyclonal
antibody contract manufacturing service, was added and incubated
for 30 min on ice, followed by staining with fluorescein
isothiocyanate-labeled anti-rabbit IgG antibody (cat. no. ab6717;
Abcam) for 30 min on ice. The fluorescence intensities of TaKE1 for
each cell type were measured as binding properties using an Accuri
C6 flow cytometer and analyzed using BD Accuri™ C6 Software
(version 1.0.264.21; both from BD Biosciences).
In vitro growth inhibition assay
The in vitro growth inhibition of cancer
cells was examined using an MTS assay kit (CellTiter 96® AQueous
Non-Radioactive Cell Proliferation Assay; Promega Corporation), as
previously reported (24). In
brief, TFK-1 cells [5,000 cells in 100 µl of RPMI-1640 medium
(Sigma-Aldrich: Merck KGaA) supplemented with 10% fetal bovine
serum (Biowest), 100 U/ml penicillin, and 100 µg/ml streptomycin]
were plated on 96-well, half-area (A/2), flat-bottomed plates
(Corning, Inc.). Cells were cultured at 37°C overnight to allow
well adhesion. After removal of the culture medium by aspiration,
100 µl of effector cells plus various concentrations of recombinant
antibodies were added to each well. After culture of the cells for
16 h at 37°C, each well was washed with PBS three times to remove
effector cells and dead target cells, and 90 µl of culture medium
plus 10 µl of MTS reagent was added to each well. The plates were
incubated for 1 h at 37°C and then read on a microplate reader at a
wavelength of 490 nm. Growth inhibition of target cells was
calculated as follows: Percentage growth inhibition of target
cells=[1-(A490 of experiment-A490 of background)/(A490 of
control-A490 of background)] ×100.
Statistical analysis
Data are presented as the mean ± standard deviation
are representative of at least two independent experiments. All
statistical analyses were performed with EZR (version 1.61; Saitama
Medical Center, Jichi Medical University), which is a graphical
user interface for R (version 4.2.2; The R Foundation for
Statistical Computing). More precisely, it is a modified version of
R commander designed to add statistical functions frequently used
in biostatistics (26). The
significance of the results was analyzed by one-way ANOVA and
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Design of three types of TaKEs
A total of three types of TaKEs were designed with
specificity to cancer cells, T cells and NK cells: i) TaKE1, ii)
TaKE2 and iii) TaKE3 (Fig. 1A). All
TaKEs contained two Fvs against cancer and one Fv for each effector
cell. A previous study revealed that higher affinities against
cancer cells than those against effector cells are important for
reducing severe adverse effects caused by excessive activation of
effector cells (27). A
tetrabody-like configuration, formed from a circularly tetramerized
single-chain Fv (scFv) (28), was
predicted for TaKE1 and TaKE2, as short five amino acid linkers
were used as middle linkers of each chimeric single-chain component
(Fig. 1B). By contrast, a tandem
scFv-based configuration was predicted for TaKE3, as short five
amino acid linkers were used to connect each scFv to prevent
interactions between different scFvs (Fig. 1B). Two anti-EGFR antibody clones,
528 and 225, competitively target similar epitope regions (29) and show similar binding constants
(30,31). To reduce unfavorable interactions
between noncognate Fv clones through domain swapping, combinations
of 528 and 225 Fv were integrated into TaKE2 and TaKE3. To simplify
the purification procedure using protein A chromatography, all
TaKEs were prepared in a human IgG1 Fc fusion format. Additionally,
the HRV3C protease recognition site (LEVLFQGP) was inserted before
the hinge region to produce TaKEs without Fc.
Preparation and evaluation of
TaKE-Fcs
To identify functional trispecific antibodies, a
cancer growth inhibition assay using protein A-purified TaKE-Fcs
was performed. After transient expression using the Expi293
Expression System, TaKE-Fcs were purified from the culture
supernatant using protein A chromatography and separated from the
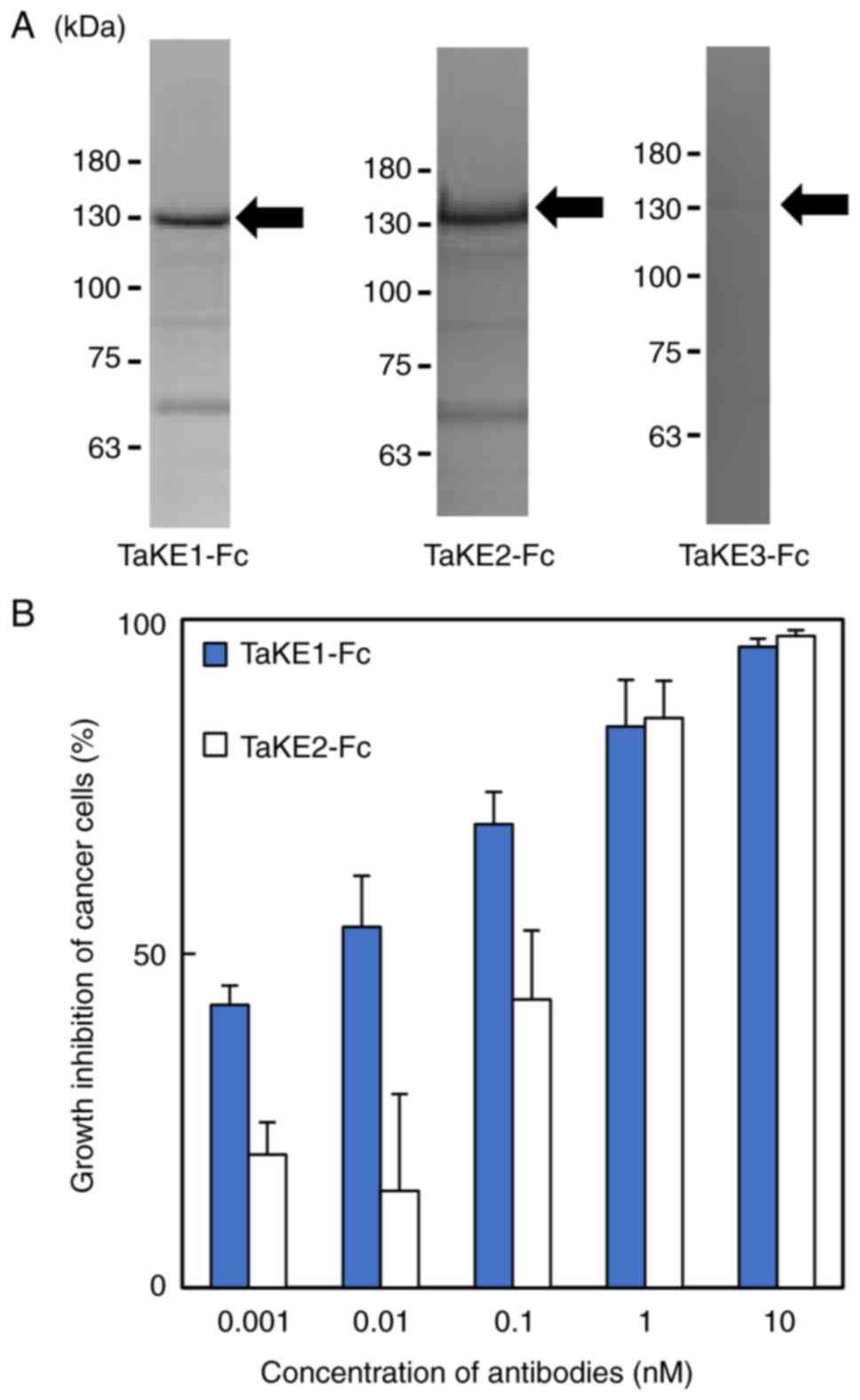
products using SDS-PAGE. Bands corresponding to the estimated
molecular weights of TaKE1-Fc and TaKE2-Fc (~130 kDa) were
observed, whereas these bands were not observed for TaKE3-Fc
(Fig. 2A). The yields were 1.7 and
0.23 mg/l for TaKE1-Fc and TaKE2-Fc, respectively. Both TaKE1-Fc
and TaKE2-Fc inhibited cancer growth in a dose-dependent manner,
although TaKE1-Fc exhibited stronger effects at lower
concentrations (Fig. 2B). These
results demonstrated that the trispecific antibodies were
successfully prepared. TaKE1-Fc exhibited high yield and activity
and was selected for further evaluation.
Preparation of TaKE1 with no Fc
region
For further functional evaluation using the highly
purified samples, TaKE1 with no Fc region was prepared because it
can bind and activate NK cells. TaKE1 was prepared from TaKE1-Fc
using HRV3C protease digestion (as described in the Materials and
methods section; Fig. 3A). After
removing the protease and undigested TaKE1-Fc, gel filtration was
performed to fractionate the TaKE1 monomer. Three peaks were
estimated to correspond to the TaKE1 dimer, TaKE1 monomer and
impurities, respectively, from the calibration protein information
(Fig. 3B). SDS-PAGE for each
fraction supported these results and revealed that TaKE1 monomer
was prepared with high purity and with the theoretical molecular
weight in the peak 2 fraction (Fig.
3C). Thus, the peak 2 fraction was used for functional
evaluation.
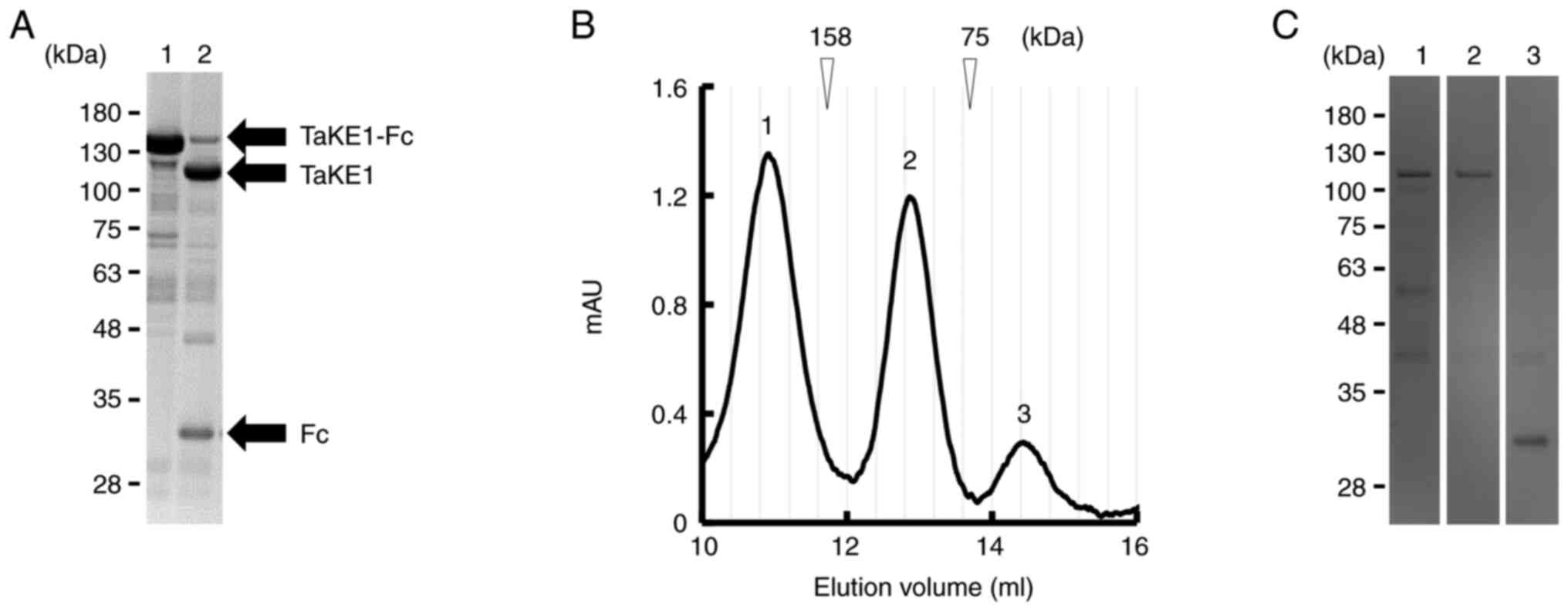 | Figure 3.Preparation of TaKE from the
Fc-fusion format. (A) Reducing SDS-PAGE analysis; lane 1, protein A
chromatography-purified TaKE1-Fc; lane 2, after HRV3C protease
digestion. (B) Gel filtration of TaKE1 after removal of HRV3C
protease by glutathione Sepharose 4B chromatography followed by
removal of Fc through protein A. The elution peaks were numbered as
1–3 (peak 1, TaKE1 dimer; peak 2, TaKE1 monomer; peak 3,
impurities). (C) Reducing SDS-PAGE for each peak after gel
filtration of TaKE1. Lanes 1–3 corresponded to the elution peaks in
the chromatograph of B (lane 1, TaKE1 dimer; lane 2, TaKE1 monomer;
lane 3, impurities). AU, absorbance unit; TaKE, T cell and natural
killer cell engager. |
Functional evaluations of TaKE1
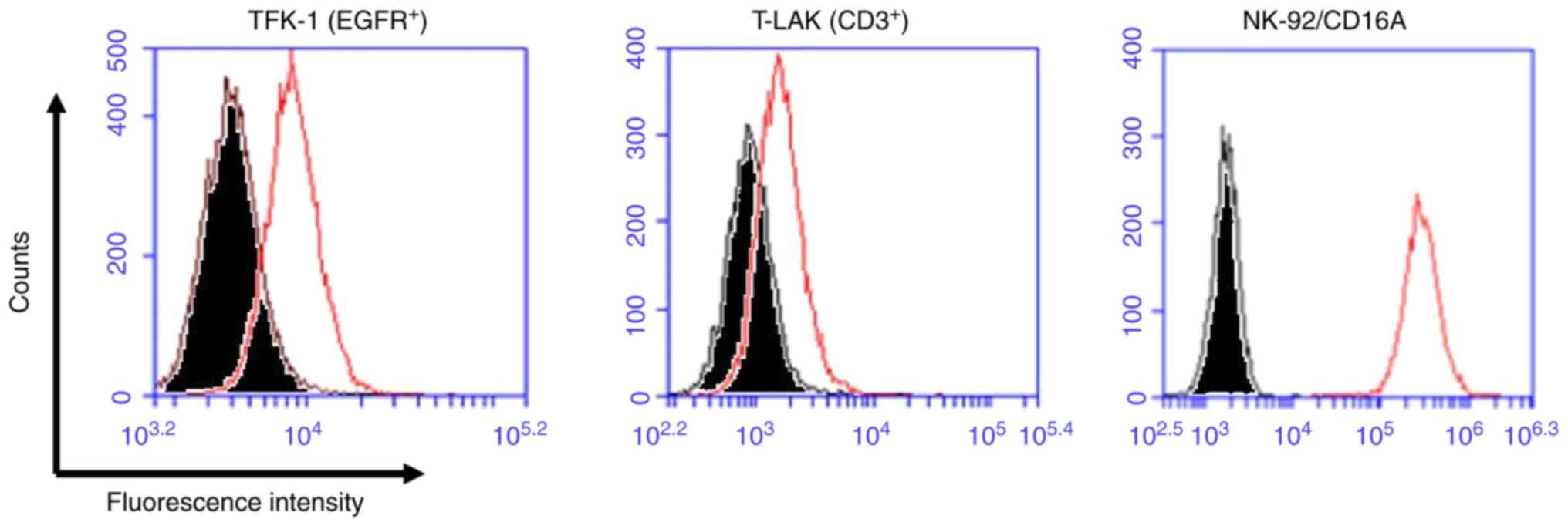
To investigate the trispecific binding abilities of
TaKE1, flow cytometric analyses using EGFR-positive TFK-1 cells,
CD3-positive T-LAK cells and CD16-positive NK92/CD16A cells were
performed. Although the shifts in the fluorescence intensity
differed for each target, with relatively low values observed for
TFK-1 cells and T-LAK cells and high values observed for NK92/CD16A
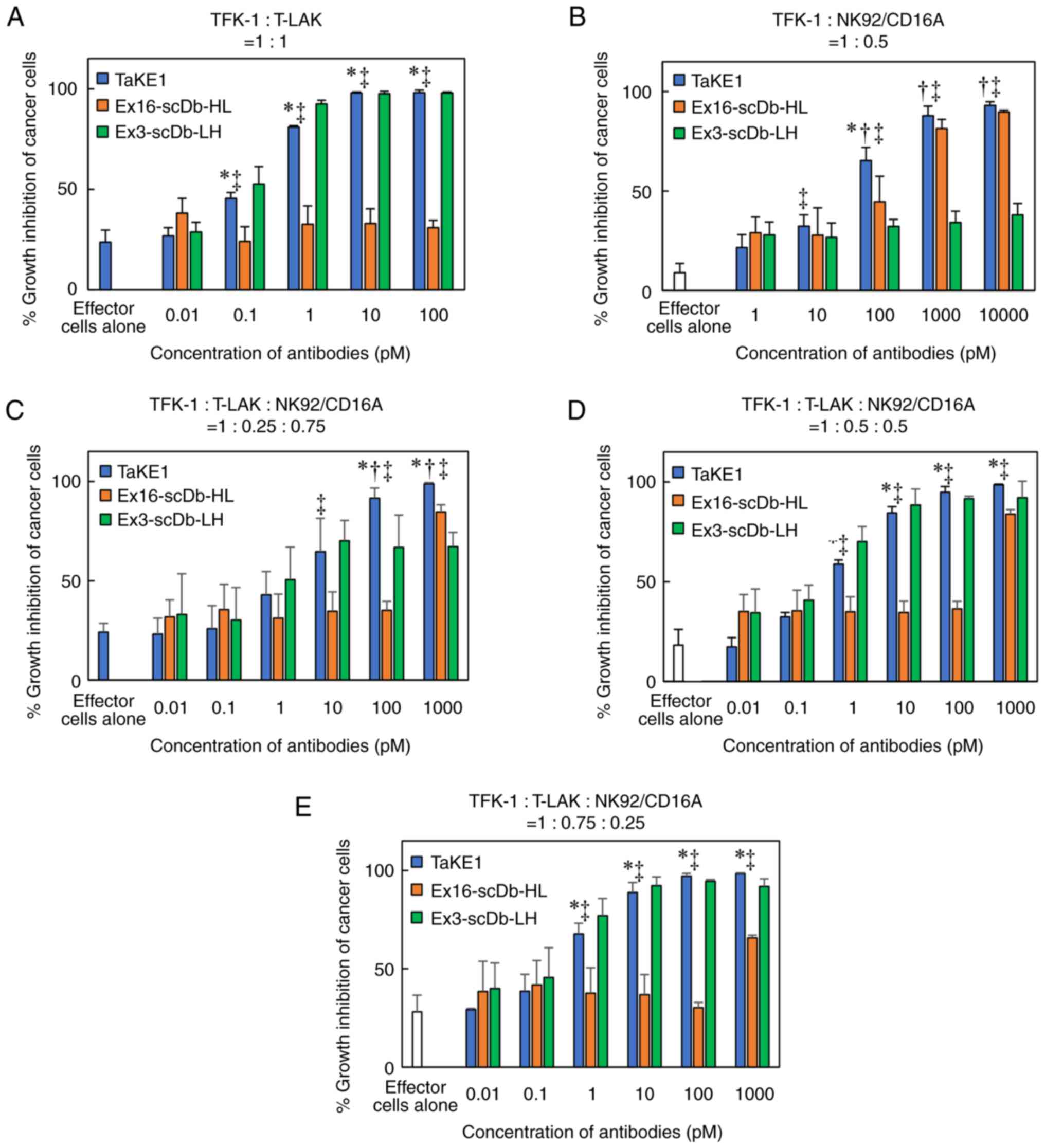
cells, the trispecificity of TaKE1 was confirmed (Fig. 4). Subsequently, the growth
inhibitory effects of TaKE1 versus those of bsAbs were evaluated in
an MTS assay using T-LAK cells or NK92/CD16A cells as effector
cells (Fig. 5A and B). The bsAbs
Ex16-scDb-HL and Ex3-scDb-LH inhibited cancer growth only when the
corresponding effector cells, which were NK92/CD16A cells for
Ex16-scDb-HL and T-LAK cells for Ex3-scDb-LH, were cocultured. By
contrast, TaKE1 revealed comparable effects with Ex16-scDb-HL in
NK92/CD16A cells and with Ex3-scDb-LH in T-LAK cells. Subsequently,
the effects when both effector cells were present at different
ratios were evaluated (Fig. 5C-E).
Ex16-scDb-HL was ineffective at concentrations below 100 pM. By
contrast, TaKE1 was effective at most concentrations and
demonstrated slightly stronger effects than those of Ex3-scDb-LH,
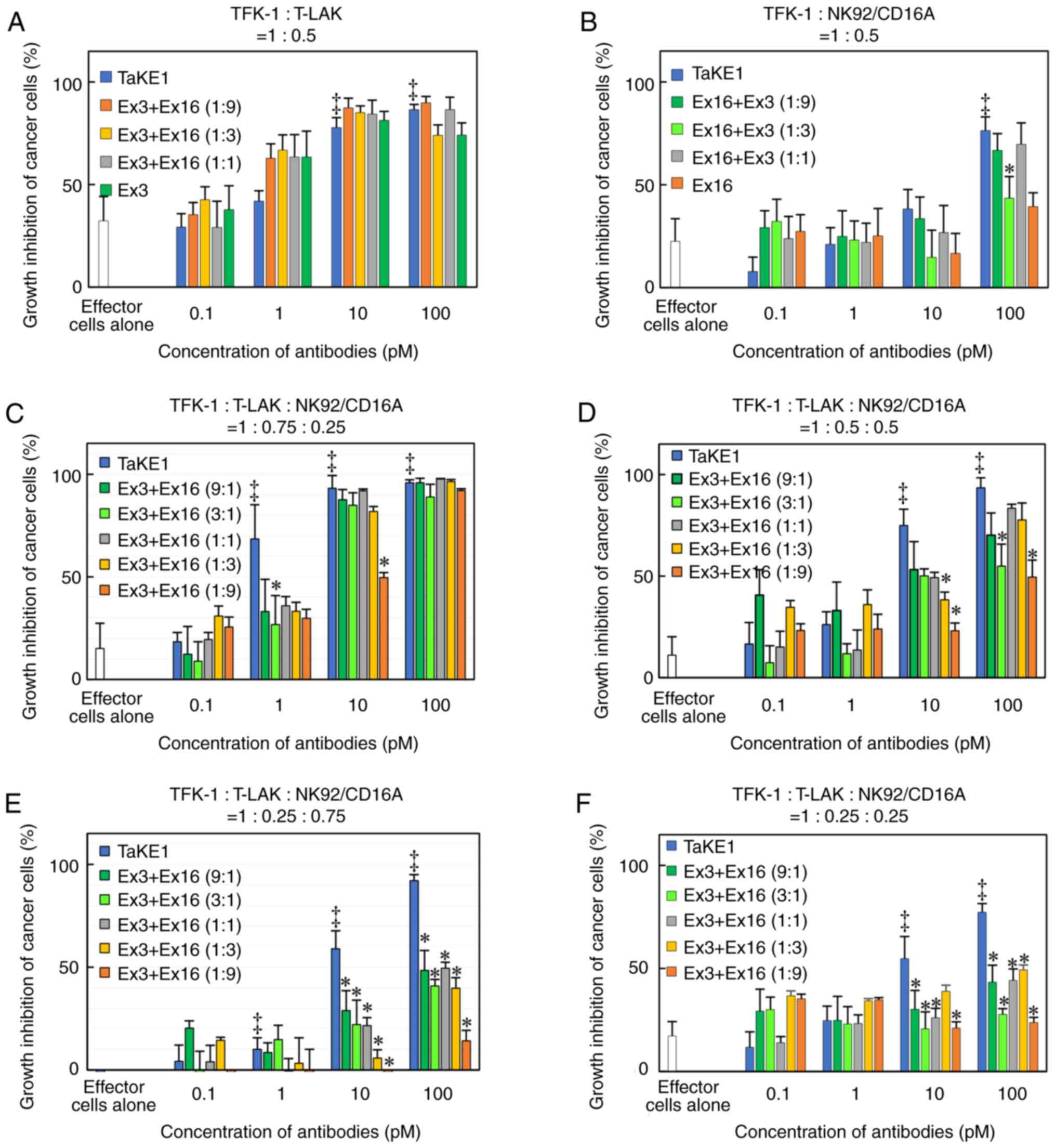
particularly at a lower ratio of T-LAK cells. Finally, the effects
of TaKE1 were compared with mixtures of Ex16-scDb-HL and
Ex3-scDb-LH, under different ratios of effector cells, to
investigate the potency of TaKE1 to the combination of two bsAbs
(Fig. 6). Under most conditions,
TaKE1 exhibited at least comparative effects as a mixture of bsAbs
and strong effects in the small population of T-LAK cells. These
results indicated the successful preparation of a trispecific
antibody that recruits T cells and NK cells, with comparable
effects as the combination of two bsAbs.
Discussion
To induce therapeutic effects independent of the
populations of T cells and NK cells among TILs, three trispecific
antibodies were designed, namely TaKEs. All TaKEs contained two Fvs
against cancer and one Fv for each effector cell for effective
tumor targeting and reducing overactivation of effector cells
(27), as well as a human Fc region
to simplify protein A purification. TaKE1-Fc and TaKE2-Fc, which
were predicted to form circular tetrabody-like configurations, were
expressed transiently in Expi293 cells (Fig. 2A), whereas TaKE3-Fc with four scFvs
connected tandemly was not expressed. In scFvs, the length and
composition of the polypeptide linker between the VH and VL domains
strongly influences formation of the multimeric structure; such
scFv dimers, trimers and tetramers are known as diabodies,
triabodies and tetrabodies, respectively (32–36).
Multimerization depends on the antibody clones. It has been
previously reported that anti-EGFR scFv and Ex3 diabodies both
include anti-EGFR Fv clone 528 and partially form stable tetrabody
(37) and tetrabody-like structures
(38), respectively. This
characteristic may have contributed to the successful preparation
of TaKE1-Fc and TaKE2-Fc, as well as the higher expression yields
and activities of TaKE1-Fc with two 528 Fvs than those of
TaKE2-Fc.
For further functional evaluation, the Fc region of
TaKE1-Fc was removed through HRV3C protease digestion because the
Fc region can bind and activate NK cells. Although complete
digestion of Fc was confirmed using SDS-PAGE (Fig. 3A), the peak corresponding to the
TaKE1 dimer revealed a similar intensity as the monomer peak
(Fig. 3B and C). Formation of
multimers by domain swapping often occurs during the development of
multispecific and/or multivalent antibodies with multiple Fv
domains, as it has been previously reported (22,39).
The length of the linkers and domain order of Fvs, which can
markedly affect the function (22,40),
are currently being optimized by the authors, to reduce the
formation of such multimers and increase the number of monomers for
further functional evaluation.
Although TaKE1 showed trispecificity with the
ability to bind TFK-1 cells, T-LAK cells, and NK92/CD16A cells
(Fig. 4), strong interactions were
clearly observed only in NK92/CD16A cells. Anti-Ex3 serum was used
as the detection antibody because TaKE1 possesses no potential
detection tags; the epitopes in TaKE1 for anti-Ex3 serum may be
hidden by binding of TaKE1 to EGFR on TFK-1 cells and CD3 on T-LAK
cells. TaKE1 was effective in the presence of either NK92/CD16A
cells or T-LAK cells, unlike bsAbs (Fig. 5A and B), supporting the
trispecificity of TaKE1. In the presence of both effector cells,
TaKE1 was more effective than Ex16-scDb-HL at most concentrations
and than Ex3-scDb-LH only at a lower ratio of T-LAK cells (Fig. 5 C-E). Although further detailed
evaluation is needed, these results indicate that TaKE1 binds
predominantly to T-LAK cells. TaKE1 also demonstrated comparable or
higher cancer growth inhibitory effects than the combination of the
two types of bsAbs (Fig. 6).
Although other in vitro studies such as cell cycle analysis
and apoptosis analysis, and in vivo studies to evaluate
antitumor effects should be performed, a functional trispecific
antibody that may recruit both T and NK cells was developed.
Various multispecific and/or multivalent antibody
formats have been designed using Fvs, as well as single-domain
antibodies, such as VHH (17,41,42).
However, these formats may not be functional and practical because
their effects are highly dependent on the therapeutic strategy,
targets and antibody clones. Especially, in the series of the
comprehensive development of bsAbs, an improvement of ~1,000-times
was identified in the cytotoxicity by screening effective
combinations of antibody clones even for the same targets (43). Antibody combinations and structural
formats may produce steric hindrance in the cross-linking of target
cells (22,44), and these may also affect the
efficacy of trispecific antibodies. In actuality, the TaKE format
with a circular tetrabody-like configuration also has antibody
clone dependencies; TaKE1 was superior to TaKE2 and further
investigation of antibody clones may result in the enhancement of
cytotoxicity. However, the highly potent cytotoxicity would raise
concerning side effects. In this case, a design of prodrug
antibodies is effective (45), and
the universal design of prodrug antibodies was recently reported by
the authors (46). One advantage of
the TaKE format without the Fc region is the possibility of
producing these antibodies using a cost-effective microbial
expression system. The authors of the present study have previously
prepared anti-EGFR tetrabody (37)
and Ex3 tetrabody (38), both of
which are similar in size to TaKE, using Escherichia coli
and reported the preparation of functional bsAbs using the
gram-positive Brevibacillus choshinensis expression system
(13). The authors are working to
prepare TaKEs using microbial expression systems.
While the development of complex multispecific
antibodies has been progressing, one limitation is methods to
evaluate and compare the efficacy of them precisely. General
methods for in vitro growth inhibition assay can not always
reflect the behavior of cancer therapeutic multispecific antibodies
in solid tumors with TILs. The use of a xenografted mouse model
with a surgical resection containing TILs from cancer patients is a
possibility (47); however, this
involves extensive challenges especially in the regulations. The
generation of spheroids formed from cancer cell lines containing
some effector cells is an ideal alternative method (48). A cancer spheroid model with T cells
and NK cells would be helpful to evaluate and compare trispecific
antibodies such as TaKEs in the future.
In the development of multispecific antibodies, the
designs to redirect some types of immune cells, especially T cells
and NK cells, toward cancer cells have been largely studied
(10,11). However, one limitation is the
differences in the abundance ratios of T cells and NK cells in
tumor cells (19) and
administration of two bsAbs recruiting T cells and NK cells is not
a realistic therapeutic strategy due to the high production costs.
Although the preparation of homogeneous molecules with high yields
is required for additional in vitro investigations and in
vivo experiments to strengthen the utility of the TaKE, a
functional trispecific antibody with potentially strong therapeutic
effects that are independent of the T cell and NK cell populations
was generated.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms Hiromi Ogata
(Department of Biomolecular Engineering, Graduate School of
Engineering, Tohoku University, Sendai, Japan), Ms Mika Ohta, Ms
Sayuri Murasaki and Ms Maiko Ueki (all from Department of
Biotechnology and Life Science, Graduate School of Engineering,
Tokyo University of Agriculture and Technology, Tokyo, Japan) for
their excellent technical assistance.
Funding
The present research was funded by Grants-in-Aid for Scientific
Research from the Japan Society for the Promotion of Science (JSPS)
(grant nos. 21K18321 and 22H02915).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
RA and IK conceived the study. RA, TN and IK
designed the study. KK, AK and SS conducted the experiments. RA and
KK analyzed the data and confirm the authenticity of all the raw
data. KK and RA wrote the manuscript. TN and IK provided conceptual
advice for the study. RA and KK edited the manuscript. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no. R2-60)
by the Biosafety Subcommittee for Safe Handling of Living Modified
Organisms at Tokyo University of Agriculture and Technology (Tokyo,
Japan) and carried out according to the guidelines of the
committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NK
|
natural killer
|
|
TaKE
|
T cell and NK cell engager
|
|
bsAbs
|
bispecific antibodies
|
|
EGFR
|
epidermal growth factor receptor
|
|
TILs
|
tumor-infiltrating lymphocytes
|
|
Fv
|
variable fragment
|
|
scFv
|
single-chain Fv
|
|
SDS-PAGE
|
sodium dodecyl sulfate-polyacrylamide
gel electrophoresis
|
|
T-LAK
|
lymphokine-activated killer cells with
T-cell phenotype
|
References
|
1
|
Kaplon H and Reichert JM: Antibodies to
watch in 2018. MAbs. 10:183–203. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ecker DM, Jones SD and Levine HL: The
therapeutic monoclonal antibody market. MAbs. 7:9–14. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lou H and Cao X: Antibody variable region
engineering for improving cancer immunotherapy. Cancer Commun
(Lond). 42:804–827. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arlotta KJ and Owen SC: Antibody and
antibody derivatives as cancer therapeutics. Wiley Interdiscip Rev
Nanomed Nanobiotechnol. 11:e15562019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nunez-Prado N, Compte M, Harwood S,
Álvarez-Méndez A, Lykkemark S, Sanz L and Álvarez-Vallina L: The
coming of age of engineered multivalent antibodies. Drug Discov
Today. 20:588–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jaiswal D, Verma S, Nair DT and Salunke
DM: Antibody multispecificity: A necessary evil? Mol Immunol.
152:153–161. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Steinhardt JJ, Guenaga J, Turner HL, McKee
K, Louder MK, O'Dell S, Chiang CI, Lei L, Galkin A, Andrianov AK,
et al: Rational design of a trispecific antibody targeting the
HIV-1 Env with elevated anti-viral activity. Nat Commun. 9:8772018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong X and D'Antona AM: Recent advances
in the molecular design and applications of multispecific
biotherapeutics. Antibodies (Basel). 10:132021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Castoldi R, Jucknischke U, Pradel LP,
Arnold E, Klein C, Scheiblich S, Niederfellner G and Sustmann C:
Molecular characterization of novel trispecific ErbB-cMet-IGF1R
antibodies and their antigen-binding properties. Protein Eng Des
Sel. 25:551–559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kamakura D, Asano R and Yasunaga M: T cell
bispecific antibodies: An antibody-based delivery system for
inducing antitumor immunity. Pharmaceuticals (Basel). 14:11722021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ferrini S, Cambiaggi A, Cantoni C,
Canevari S, Mezzanzanica D, Colnaghi MI and Moretta L: Targeting of
T or NK lymphocytes against tumor cells by bispecific monoclonal
antibodies: Role of different triggering molecules. Int J Cancer
Suppl. 7:15–18. 1992.PubMed/NCBI
|
|
12
|
Asano R, Sone Y, Makabe K, Tsumoto K,
Hayashi H, Katayose Y, Unno M, Kudo T and Kumagai I: Humanization
of the bispecific epidermal growth factor receptor × CD3 diabody
and its efficacy as a potential clinical reagent. Clin Cancer Res.
12:4036–4042. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Asano R, Kuroki Y, Honma S, Akabane M,
Watanabe S, Mayuzumi S, Hiyamuta S, Kumagai I and Sode K:
Comprehensive study of domain rearrangements of single-chain
bispecific antibodies to determine the best combination of
configurations and microbial host cells. MAbs. 10:854–863. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Asano R, Nakayama M, Kawaguchi H, Kubota
T, Nakanishi T, Umetsu M, Hayashi H, Katayose Y, Unno M, Kudo T and
Kumagai I: Construction and humanization of a functional bispecific
EGFR CD16 diabody using a refolding system. FEBS J. 279:223–233.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuwahara A, Nagai K, Nakanishi T, Kumagai
I and Asano R: Functional domain order of an anti-EGFR × Anti-CD16
bispecific diabody involving NK cell activation. Int J Mol Sci.
21:89142020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meermeier EW, Welsh SJ, Sharik ME, Du MT,
Garbitt VM, Riggs DL, Shi CX, Stein CK, Bergsagel M, Chau B, et al:
Tumor burden limits bispecific antibody efficacy through T cell
exhaustion averted by concurrent cytotoxic therapy. Blood Cancer
Discov. 2:354–369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Beha N, Harder M, Ring S, Kontermann RE
and Müller D: IL15-based trifunctional antibody-fusion proteins
with costimulatory TNF-superfamily ligands in the single-chain
format for cancer immunotherapy. Mol Cancer Ther. 18:1278–1288.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu L, Seung E, Xu L, Rao E, Lord DM, Wei
RR, Cortez-Retamozo V, Ospina B, Posternak V, Ulinski G, et al:
Trispecific antibodies enhance the therapeutic efficacy of
tumor-directed T cells through T cell receptor co-stimulation. Nat
Cancer. 1:86–98. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geissler K, Fornara P, Lautenschlager C,
Holzhausen HJ, Seliger B and Riemann D: Immune signature of tumor
infiltrating immune cells in renal cancer. Oncoimmunology.
4:e9850822015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Asano R, Nagai K, Makabe K, Takahashi K,
Kumagai T, Kawaguchi H, Ogata H, Arai K, Umetsu M and Kumagai I:
Structural considerations for functional anti-EGFR × anti-CD3
bispecific diabodies in light of domain order and binding affinity.
Oncotarget. 9:13884–13893. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Miyazaki J, Takaki S, Araki K, Tashiro F,
Tominaga A, Takatsu K and Yamamura K: Expression vector system
based on the chicken beta-actin promoter directs efficient
production of interleukin-5. Gene. 79:269–277. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Asano R, Shimomura I, Konno S, Ito A,
Masakari Y, Orimo R, Taki S, Arai K, Ogata H, Okada M, et al:
Rearranging the domain order of a diabody-based IgG-like bispecific
antibody enhances its antitumor activity and improves its
degradation resistance and pharmacokinetics. MAbs. 6:1243–1254.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saijyo S, Kudo T, Suzuki M, Katayose Y,
Shinoda M, Muto T, Fukuhara K, Suzuki T and Matsuno S:
Establishment of a new extrahepatic bile duct carcinoma cell line,
TFK-1. Tohoku J Exp Med. 177:61–71. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Asano R, Watanabe Y, Kawaguchi H, Fukazawa
H, Nakanishi T, Umetsu M, Hayashi H, Katayose Y, Unno M, Kudo T and
Kumagai I: Highly effective recombinant format of a humanized
IgG-like bispecific antibody for cancer immunotherapy with
retargeting of lymphocytes to tumor cells. J Biol Chem.
282:27659–27665. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakadate Y, Kodera Y, Kitamura Y,
Shirasawa S, Tachibana T, Tamura T and Koizumi F: KRAS mutation
confers resistance to antibody-dependent cellular cytotoxicity of
cetuximab against human colorectal cancer cells. Int J Cancer.
134:2146–2155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie Z, Shi M, Feng J, Yu M, Sun Y, Shen B
and Guo N: A trivalent anti-erbB2/anti-CD16 bispecific antibody
retargeting NK cells against human breast cancer cells. Biochem
Biophys Res Commun. 311:307–312. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gall FL, Kipriyanov SM, Moldenhauer G and
Little M: Di-, tri- and tetrameric single chain Fv antibody
fragments against human CD19: Effect of valency on cell binding.
FEBS Lett. 453:164–168. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sato JD, Kawamoto T, Le AD, Mendelsohn J,
Polikoff J and Sato GH: Biological effects in vitro of monoclonal
antibodies to human epidermal growth factor receptors. Mol Biol
Med. 1:511–529. 1983.PubMed/NCBI
|
|
30
|
Makabe K, Nakanishi T, Tsumoto K, Tanaka
Y, Kondo H, Umetsu M, Sone Y, Asano R and Kumagai I: Thermodynamic
consequences of mutations in vernier zone residues of a humanized
anti-human epidermal growth factor receptor murine antibody, 528. J
Biol Chem. 283:1156–1166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li SQ, Schmitz KR, Jeffrey PD, Wiltzius
JJW, Kussie P and Ferguson KM: Structural basis for inhibition of
the epidermal growth factor receptor by cetuximab. Cancer Cell.
7:301–311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pei XY, Holliger P, Murzin AG and Williams
RL: The 2.0-A resolution crystal structure of a trimeric antibody
fragment with noncognate VH-VL domain pairs shows a rearrangement
of VH CDR3. Proc Natl Acad Sci USA. 94:9637–9642. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dolezal O, Pearce LA, Lawrence LJ, McCoy
AJ, Hudson PJ and Kortt AA: ScFv multimers of the
anti-neuraminidase antibody NC10: Shortening of the linker in
single-chain Fv fragment assembled in V(L) to V(H) orientation
drives the formation of dimers, trimers, tetramers and higher
molecular mass multimers. Protein Eng. 13:565–574. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Power BE, Doughty L, Shapira DR, Burns JE,
Bayly AM, Caine JM, Liu Z, Scott AM, Hudson PJ and Kortt AA:
Noncovalent scFv multimers of tumor-targeting anti-Lewis(y) hu3S193
humanized antibody. Protein Sci. 12:734–747. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Le Gall F, Reusch U, Moldenhauer G, Little
M and Kipriyanov SM: Immunosuppressive properties of anti-CD3
single-chain Fv and diabody. J Immunol Methods. 285:111–127. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hudson PJ and Kortt AA: High avidity scFv
multimers; diabodies and triabodies. J Immunol Methods.
231:177–189. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Asano R, Koyama N, Hagiwara Y, Masakari Y,
Orimo R, Arai K, Ogata H, Furumoto S, Umetsu M and Kumagai I:
Anti-EGFR scFv tetramer (tetrabody) with a stable monodisperse
structure, strong anticancer effect, and a long in viFvo half-life.
FEBS Open Bio. 6:594–602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Asano R, Ikoma K, Sone Y, Kawaguchi H,
Taki S, Hayashi H, Nakanishi T, Umetsu M, Katayose Y, Unno M, et
al: Highly enhanced cytotoxicity of a dimeric bispecific diabody,
the hEx3 tetrabody. J Biol Chem. 285:20844–20849. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Asano R, Ikoma K, Shimomura I, Taki S,
Nakanishi T, Umetsu M and Kumagai I: Cytotoxic enhancement of a
bispecific diabody by format conversion to tandem single-chain
variable fragment (taFv): The case of the hEx3 diabody. J Biol
Chem. 286:1812–1818. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Asano R, Kumagai T, Nagai K, Taki S,
Shimomura I, Arai K, Ogata H, Okada M, Hayasaka F, Sanada H, et al:
Domain order of a bispecific diabody dramatically enhances its
antitumor activity beyond structural format conversion: The case of
the hEx3 diabody. Protein Eng Des Sel. 26:359–367. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Kang G, Yuan H, Cao X, Huang H and
de Marco A: Research progress and applications of multivalent,
multispecific and modified nanobodies for disease treatment. Front
Immunol. 12:8380822021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu X and Demarest SJ: Building blocks for
bispecific and trispecific antibodies. Methods. 154:3–9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sugiyama A, Umetsu M, Nakazawa H, Niide T,
Onodera T, Hosokawa K, Hattori S, Asano R and Kumagai I: A semi
high-throughput method for screening small bispecific antibodies
with high cytotoxicity. Sci Rep. 7:28622017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maejima A, Ishibashi K, Kim H, Kumagai I
and Asano R: Evaluation of intercellular cross-linking abilities
correlated with cytotoxicities of bispecific antibodies with domain
rearrangements using AFM force-sensing. Biosens Bioelectron.
178:1130372021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lucchi R, Bentanachs J and Oller-Salvia B:
The masking game: Design of activatable antibodies and mimetics for
selective therapeutics and cell control. ACS Cent Sci. 7:724–738.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Maejima A, Suzuki S, Makabe K, Kumagai I
and Asano R: Incorporation of a repeated polypeptide sequence in
therapeutic antibodies as a universal masking procedure: A case
study of T cell-engaging bispecific antibodies. N Biotechnol.
77:80–89. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schlereth B, Fichtner I, Lorenczewski G,
Kleindienst P, Brischwein K, da Silva A, Kufer P, Lutterbuese R,
Junghahn I, Kasimir-Bauer S, et al: Eradication of tumors from a
human colon cancer cell line and from ovarian cancer metastases in
immunodeficient mice by a single-chain Ep-CAM-/CD3-bispecific
antibody construct. Cancer Res. 65:2882–2889. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fujii H, Tanaka Y, Nakazawa H, Sugiyama A,
Manabe N, Shinoda A, Shimizu N, Hattori T, Hosokawa K, Sujino T, et
al: Compact seahorse-shaped T cell-activating antibody for cancer
therapy. Adv Ther. 1:17000312018. View Article : Google Scholar
|