Colorectal cancer (CRC) is one of the most common
malignant tumors worldwide. In China, the incidence and mortality
of CRC are increasing every year (1). Surgical resection is the mainstay of
potentially curative treatments for CRC; however, prognosis is
generally poor due to locoregional recurrence with resection alone
(2).
Cytotoxic chemotherapy is another mainstay of
treatment for CRC patients. Oxaliplatin (OXA) is a third-generation
chemotherapy drug of the diamino-cyclohexane platinum family
(3,4). Due to potent in vitro
cytotoxicity and in vivo antitumor activity, OXA-containing
regimens are effectively used as first-line chemotherapy in CRC.
However, de novo and acquired OXA resistance remains a major
challenge in CRC treatment (5). The
acquisition of OXA resistance in CRC is multi-factorial and
includes the following: cellular transport and detoxification
systems (copper transporters, solute carrier transporters and
ATP-binding cassette transporters), OXA-induced DNA adduct repair
and alterations in key cell death-related genes and/or tumor
suppressors (p53, Bcl-2 family and MMP7) (6–9).
Hypoxia is a common feature of the tumor
microenvironment that activates the expression of numerous genes
associated with cell growth, angiogenesis, metastasis and drug
resistance (10–13). Cells adapting to hypoxia have been
demonstrated to reduce the cytotoxicity of numerous drugs, such as
OXA and 5-fluorouracil (5-FU) (14,15).
Therefore, elucidating the underlying mechanisms of hypoxia-induced
drug resistance and developing more effective therapeutic regimens
to overcome hypoxia-induced drug resistance are clinical
priorities.
Accumulating evidence has shown that microRNAs
(miRs) play an important role in acquired drug resistance in
colorectal carcinoma (CRC). miRs can act as hypoxia sensors and
their levels are altered consistently in CRC cells (16). Nijhuis et al (16) indicated that treatment with miR-21
and miR-30d antagonists sensitized hypoxic CRC cells to 5-FU. Xu
et al (17) indicated that
hypoxia-inducible transcription factor 1α (HIF-1α)-mediated
suppression of miR-338-5p conferred OXA resistance in CRC
cells.
P53 is a stress-inducible transcription factor that
regulates numerous downstream genes, such as p21, Bax and GADD45,
to exert regulatory functions in multiple signaling processes
(18,19). TP53 mutation occurs in ~40-50% of
CRC (20). The TP53 mutation status
is closely related to the progression, drug resistance and outcome
of CRC (21,22). However, the effect of TP53 mutation
on drug resistance in CRC cells, particularly hypoxic CRC cells,
and the role of miRs during this process remain to be
elucidated.
The aim of the present study was to investigate how
p53 affects hypoxia-induced OXA resistance by regulating miR
expression in CRC.
For OXA chemotherapy, OXA (MilliporeSigma) solution
was prepared with cell culture medium and the working concentration
is 5, 10, 20, 40 and 60 µM. Cells were cultured in OXA for 24 h
before other experiments were carried out. For the hypoxic culture,
cells were cultured at 37°C in a humidified O2
(1%)/CO2 (5%)/N2 (94%) incubator (Thermo Fisher
Scientific, Inc.) for 24 h (17).
Echinomycin (1 nM; MedChemExpress) dissolved in dimethyl sulfoxide
(DMSO; MilliporeSigma) was added to cell culture medium for 24 h to
inactivate HIF-1α (24).
For plasmid transfection, HCT116 and HT29 cells were
seeded in six-well plates 24 h prior to transfection in complete
medium until they reached 40~60% confluency. Plasmid DNA was
complexed with Lipofectamine 3000 and P3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Transfection media was removed and replaced with new
media at 7 h post-transfection. miR-23a, −26a, −34a, −133a, −107a,
−205 listed in Table I and
NC-mimics were synthesized (Guangzhou RiboBio Co., Ltd.) and
transfected into cells at a concentration of 10 nmol/ml with
lipofectamine 3000. All the subsequent experimentations were
carried out at 48 h post-transfection.
Normal or transfected cells (at a density of 5,000
per well) in 100 µl complete medium were seeded in one well of
96-well plates. After culturing for 24 h, cells were treated with
OXA and/or hypoxia for another 24 h, and 20 µl of the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
reagent (MTT; Beyotime Biotechnology) was added to each well and
incubated at 37°C for 4 h. After removing the medium, the blue
formazan was dissolved with 200 µl DMSO, and absorbance at 550 nm
was measured. The cellular growth inhibition rate was defined as
(1-OD550 of the experimental group)/OD550 of
the control group ×100%.
Total RNA of the OXA or hypoxia treated HCT116 cells
was isolated with RNAzol (Sigma-Aldrich; Merck KGaA). A total of 31
miRs were selected and examined to analyze the effect of miRs on
cellular chemoresistance (15 miRs) or cellular response to hypoxia
(16 miRs).
RT-qPCR was conducted to detect the enrichment of
relevant miRs. RNA was reverse transcribed to cDNA using Mir-X
miRNA First-Strand Synthesis kit according to the manufacturer's
instructionσ (Takara Biotechnology Co., Ltd.). Mir-X TB Green
RT-qPCR kit (Takara Biotechnology Co., Ltd.) was used to conduct
RT-qPCR reaction according to the manufacturer's instruction. U6
snRNA expression was used as endogenous control. The 5′ primer of
U6 is CGCTTCGGCAGCACATATAC. PCR was performed on an ABI 7500 qPCR
instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The thermocycling conditions consisted of 10 sec at 95°C followed
by 40 cycles at 95°C for 5 sec and 60°C for 35 sec. The abundance
of miR was calculated using the formula of 2−ΔΔCq
(26). The primer sequences for
amplification of miRs are listed in Table I.
Cells were lysed with lysis buffer (Beijing Solarbio
Science & Technology Co., Ltd.) complemented with protease
inhibitor cocktail (Sigma-Aldrich; Merck KGaA). Cell lysates were
centrifuged at 12,000 × g at 4°C for 20 min to remove cell debris
and insoluble material. Protein concentration was quantitated using
the Bradford protein assay kit (Beyotime Institute of
Biotechnology). Equal amounts of lysate (25 µg) were loaded per
lane and proteins resolved by 5–10% SDS-PAGE gel, semi-dry
transferred to 0.45-µm polyvinylidene fluoride membranes (EMD
Millipore). The membranes were incubated in 5% skim milk in
Tris-buffered saline Tween-20 (TBST; 10 mM Tris-Base, 150 mM NaCl,
0.05% Tween-20; pH 7.4) for 1 h at room temperature, followed by
incubation with primary antibody in 5% skim milk in TBST at 4°C
overnight. The membranes were then washed for 3×5 min in TBST, and
then incubated in TBST-diluted secondary antibodies for 45 min at
room temperature, followed by another 3×5 min washes with TBST.
Protein-antibody binding was detected with ECL Western Blotting
Substrate (Beijing Solarbio Science & Technology Co., Ltd.)
followed by exposure of the membranes to X-ray film (Kodak).
Protein expression levels were determined semiquantitatively by
densitometric analysis with the Quantity One software (version
4.6.2; Bio-Rad Laboratories, Inc.). The following antibodies were
used: anti-p53 (1:1,000; cat. no. sc-47698; Santa Cruz
Biotechnology, Inc.), anti-BCL-2 (1:1,000; cat. no. 15071),
anti-MCL-1 (1:1,000; cat. no. 39224), anti-EZH2 (1:1,000; cat. no.
5246), anti-STAT3 (1:1,000; cat. no. 9139), anti-SMAD4 (1:1,000;
cat. no. 46535; all from Cell Signaling Technology, Inc.),
anti-β-ACTIN (1:2,000; cat. no. sc-8432) and HRP-conjugated
secondary antibody (1:3,000; cat. no. sc-2357; both from Santa Cruz
Biotechnology, Inc.).
Statistical analysis was performed using SPSS
software (version 19.0; IBM Corp.). Each experiment was repeated at
least three times. Statistical significance was assessed by
comparing the mean ± SD using an unpaired Student's t-test or ANOVA
test followed by Fisher's Least Significant Difference, Bonferroni
or Sidak post-hoc tests. *P<0.05 was considered to indicate a
statistically significant difference.
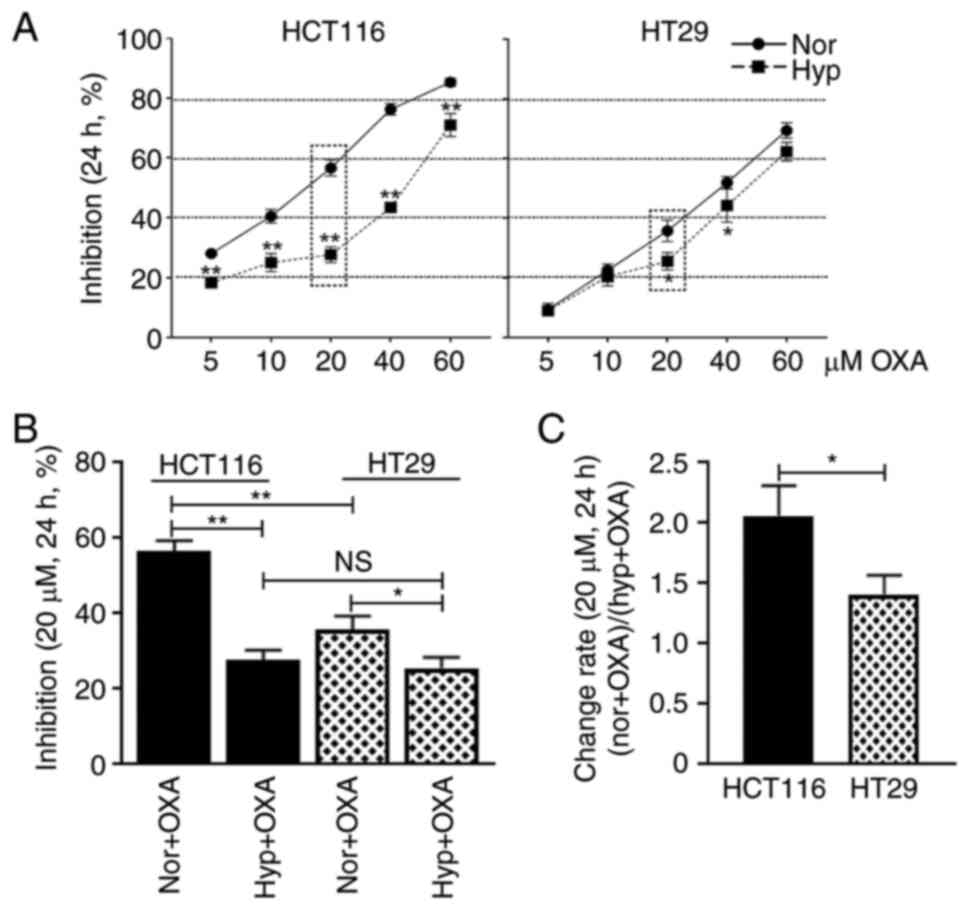
The response of HCT116 and HT29 cells to OXA was
investigated under both normoxia and hypoxia. Both cell lines were
treated with 5, 10, 20, 40 and 60 µM OXA and it was found that the
cellular growth inhibition rates increased with the increase of
drug concentration (Fig. 1A).
Taking the 20 µM OXA treatment group as an example, the cellular
growth inhibition rate of HCT116 cells was 56.6% under normoxic
condition and 27.7% under hypoxic condition (Fig. 1B; P<0.01), while the cellular
growth inhibition rate of HT29 cells was 35.7% under normoxic
condition and 25.5% under hypoxic condition (Fig. 1B; P<0.05). The cellular growth
change rate of HCT116 cells was 2.06 and of HT29 cells was 1.41
(Fig. 1C; P<0.05). These data
suggested that HCT116 cells were more sensitive to OXA than HT29
cells under normoxic condition. Additionally, although hypoxia
decreases the OXA sensitivity, it generated greater impact on
HCT116 cells than HT29 cells.
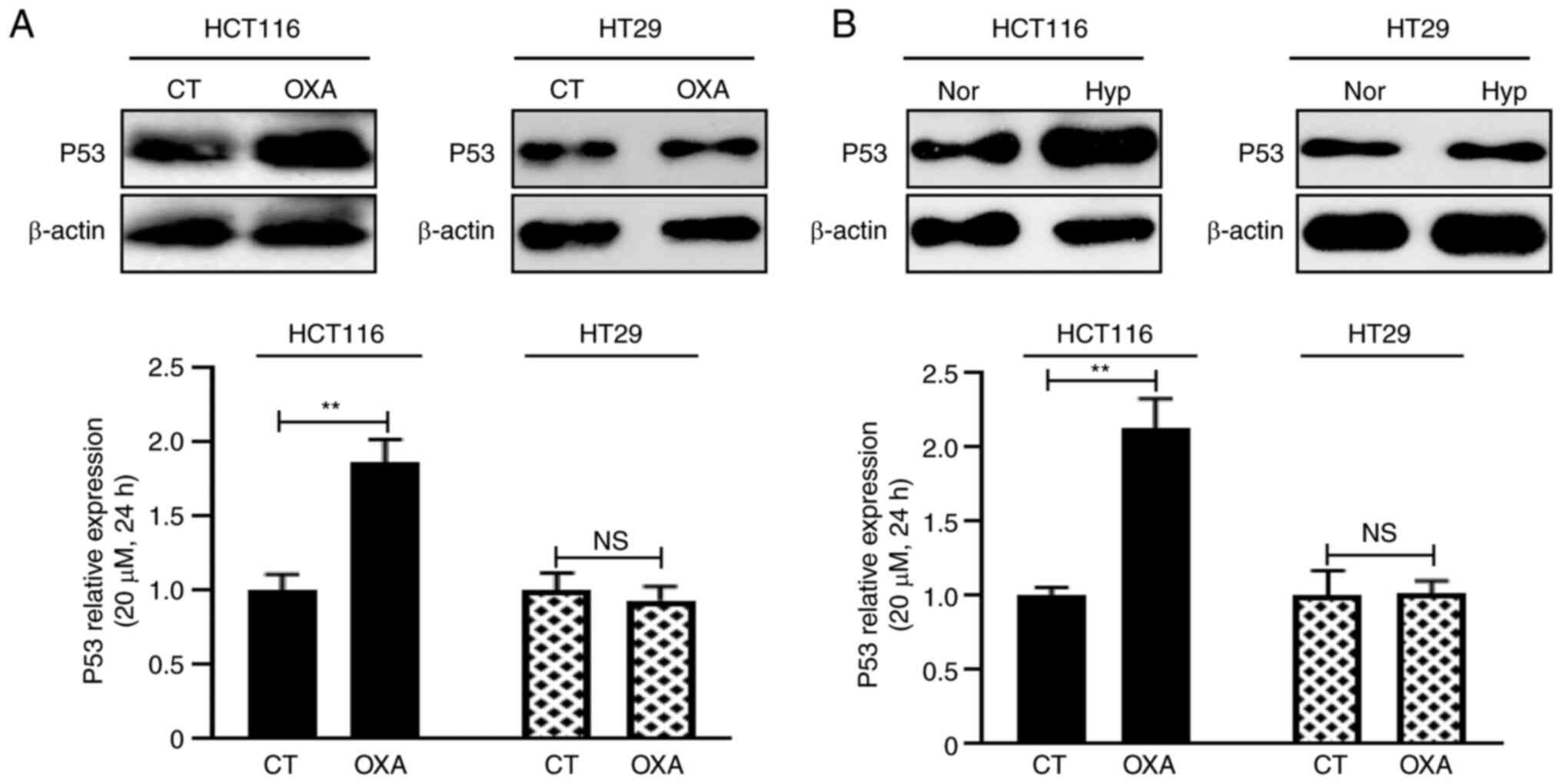
TP53 status is the typical difference between HCT116
cells and HT29 cells. P53 expression was detected after both cell
lines were treated with OXA and hypoxia and it was found that the
P53 expression level of HCT116 cells increased significantly and of
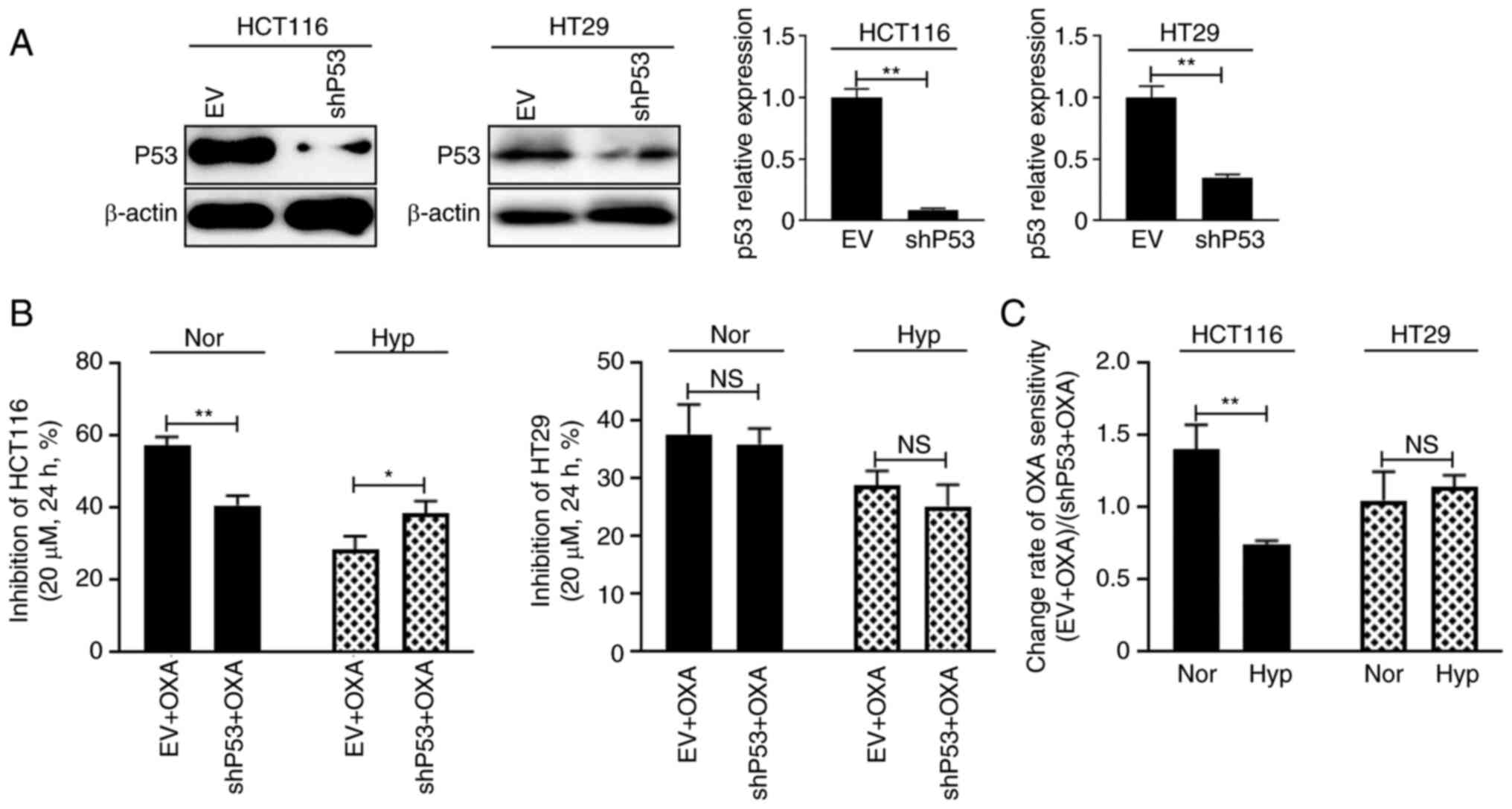
HT29 cells did not change considerably (Fig. 2A and B). P53 was then knocked down
and cell drug sensitivity to 20 µM OXA was measured (Fig. 3A). Under normoxic condition, the
cellular growth inhibition rate of HCT116 cells was 57.3% in the EV
group and 40.8% in the shP53 group (Fig. 3B, left panel; P<0.01), while
under hypoxic condition, the cellular growth inhibition rate of
HCT116 cells was 28.6% in the EV group and 38.4% in the shP53 group
(Fig. 3B, left panel; P<0.05).
These data suggested that P53WT plays different roles in
HCT116 cell drug sensitivity, enhancing drug sensitivity under
normoxic conditions but reducing drug sensitivity under hypoxic
conditions. However, knocking down TP53 did not change HT29 cell
response to OXA under either normoxia or hypoxia (Fig. 3B, right panel). The growth
inhibition change rate also reflected this phenomenon (Fig. 3C).
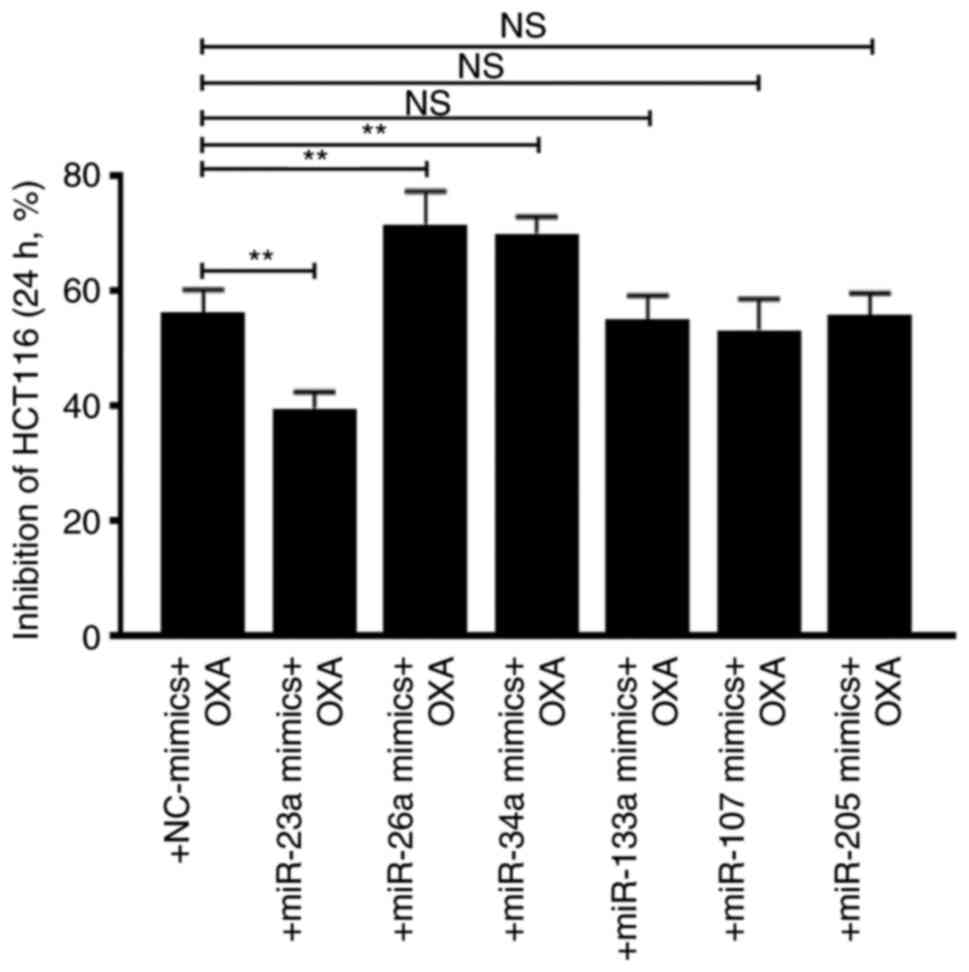
The effects of these P53-related miRs were analyzed
by overexpressing them in HCT116 cells. Cellular growth inhibition
assay showed that miR-23a decreased OXA sensitivity, whereas
miR-26a and miR-34a increased OXA sensitivity. However, miR-133a,
107 and 205 did not affect cell drug sensitivity (Fig. 5).
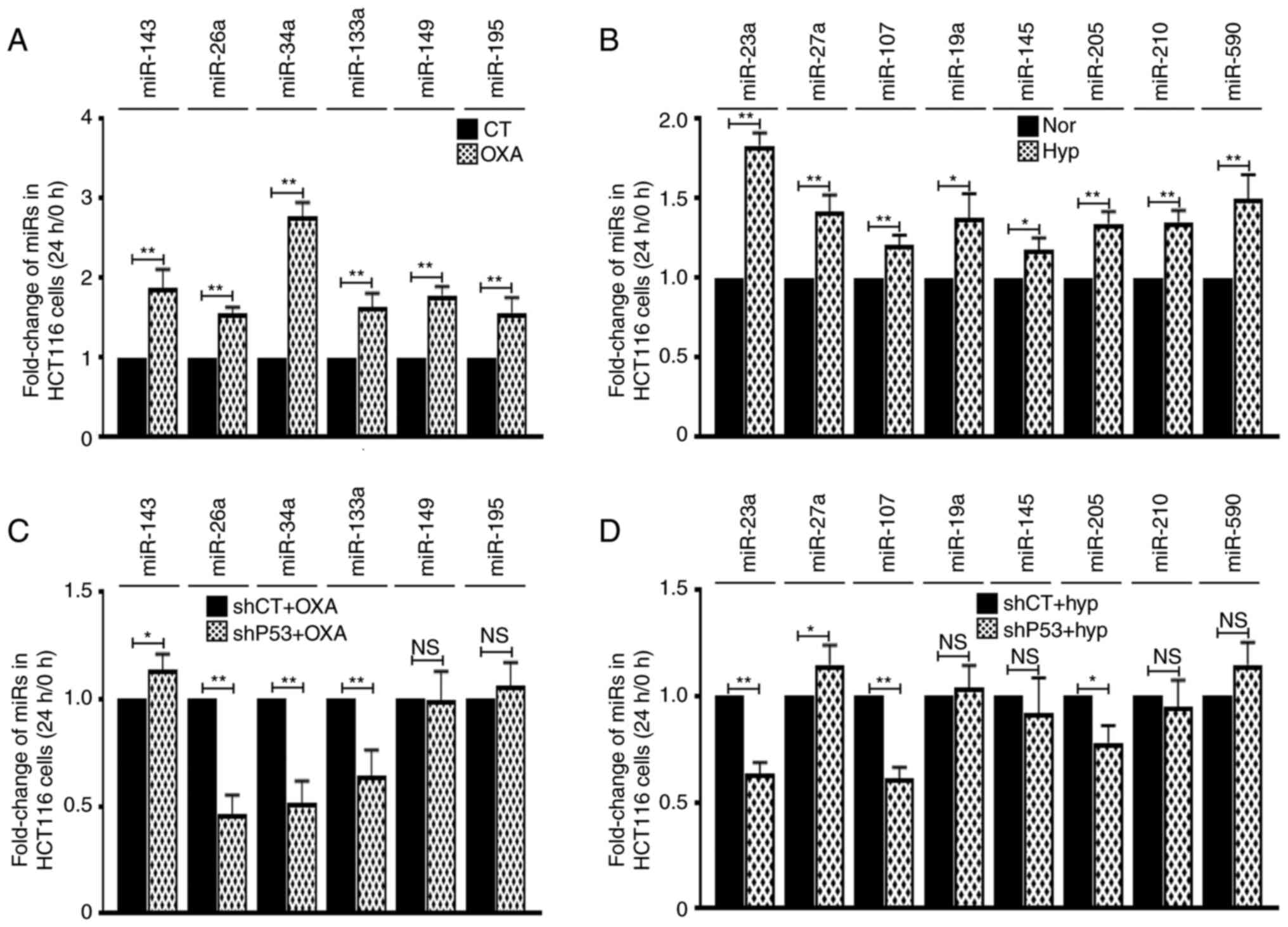
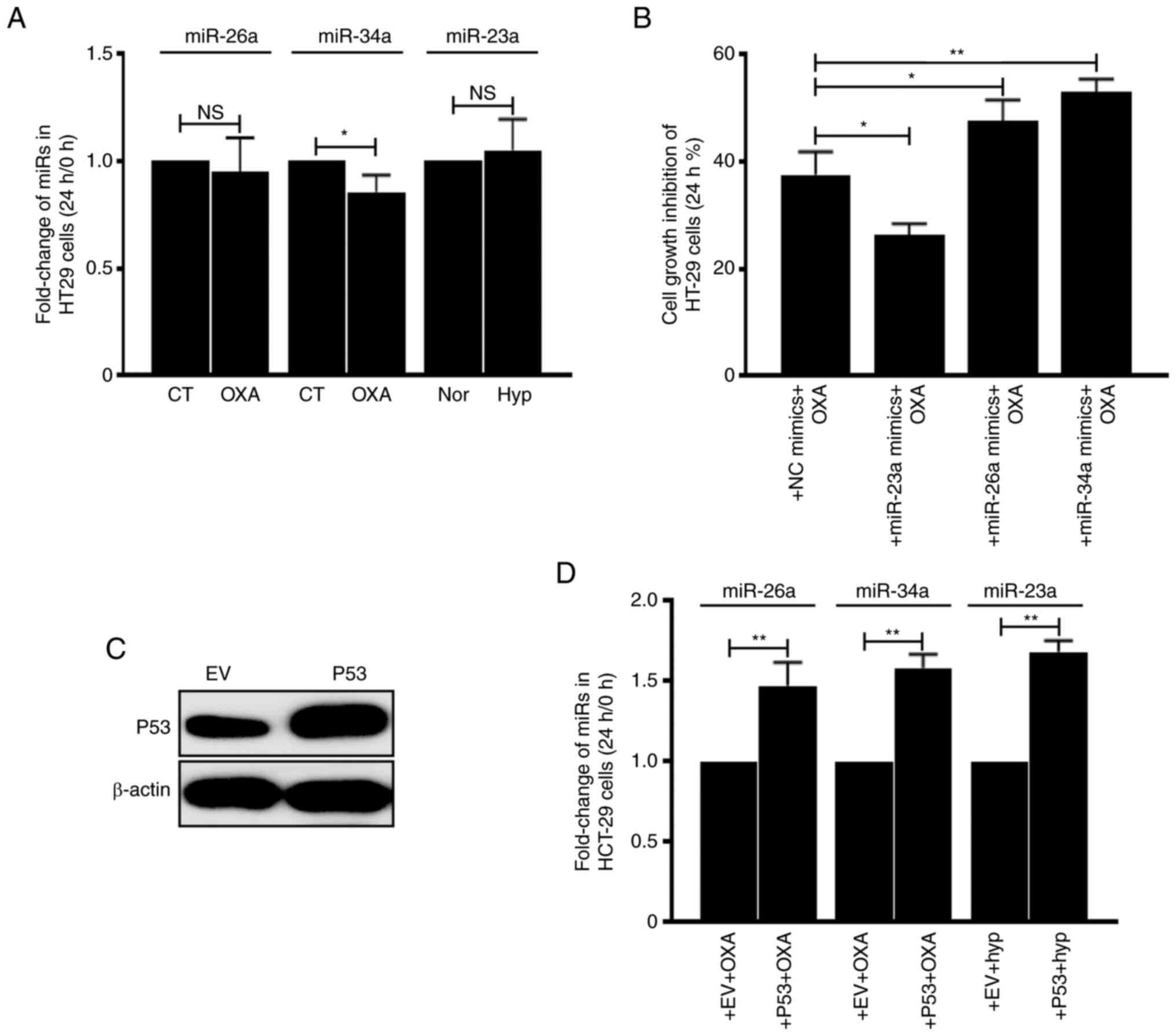
Then the association between miRs and P53 status was
analyzed. The expression levels of miRs in HT29 cells
(TP53MT) were first detected and it was identified that
OXA- or hypoxia-treatment did not upregulate miR-26a, 34a and 23a
(Fig. 6A). However, transfection of
miR-23a decreased cellular growth inhibition rate and transfection
of miR-26a and 34a increased cellular growth inhibition rate, which
was similar as they were functioning in HCT116 cells (Fig. 6B). Notably, after introducing
exogenous P53WT to HT29 cells (Fig. 6C), it was revealed that miR-26a, 34a
and 23a can be induced by OXA or hypoxia treatment (Fig. 6D). These data suggested that the
effect of these 3 miRs on CRC cell drug sensitivity depends on
P53WT.
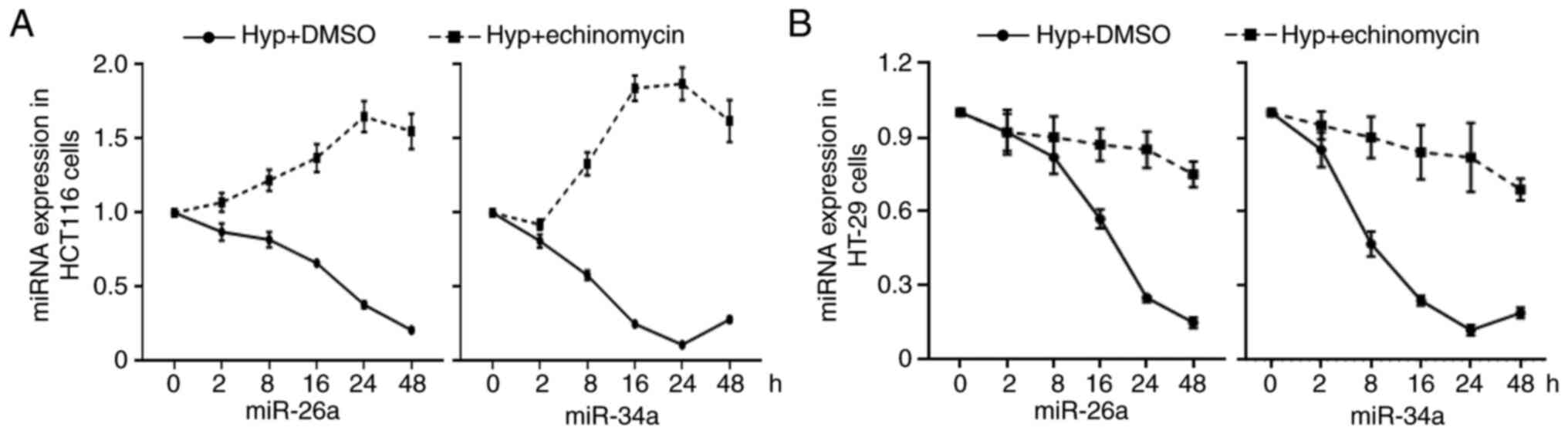
It can be observed from the aforementioned
experiments that miR-23a, 26a and 34a are all driven by P53.
However, it was strange that with hypoxia upregulating the level of
P53, it only induced the expression of miR-23a instead of affecting
the level of miR-26a and miR-34a. To address this issue, the level
of miR-26a and miR-34a was examined after CRC cells were treated
with hypoxia for 2, 8, 16, 24 and 48 h. It was demonstrated that
hypoxia significantly suppressed the expression levels of these two
miRs in both HCT116 and HT29 cells (Fig. 7A and B). However, administration of
HIF-1α inhibitor echinomycin reversed the inhibition effect of
hypoxia on miRs in HCT116 cells but not in HT29 cells (Fig. 7A and B). These data suggested that
hypoxia and P53WT synergistically altered expression of
miRs.
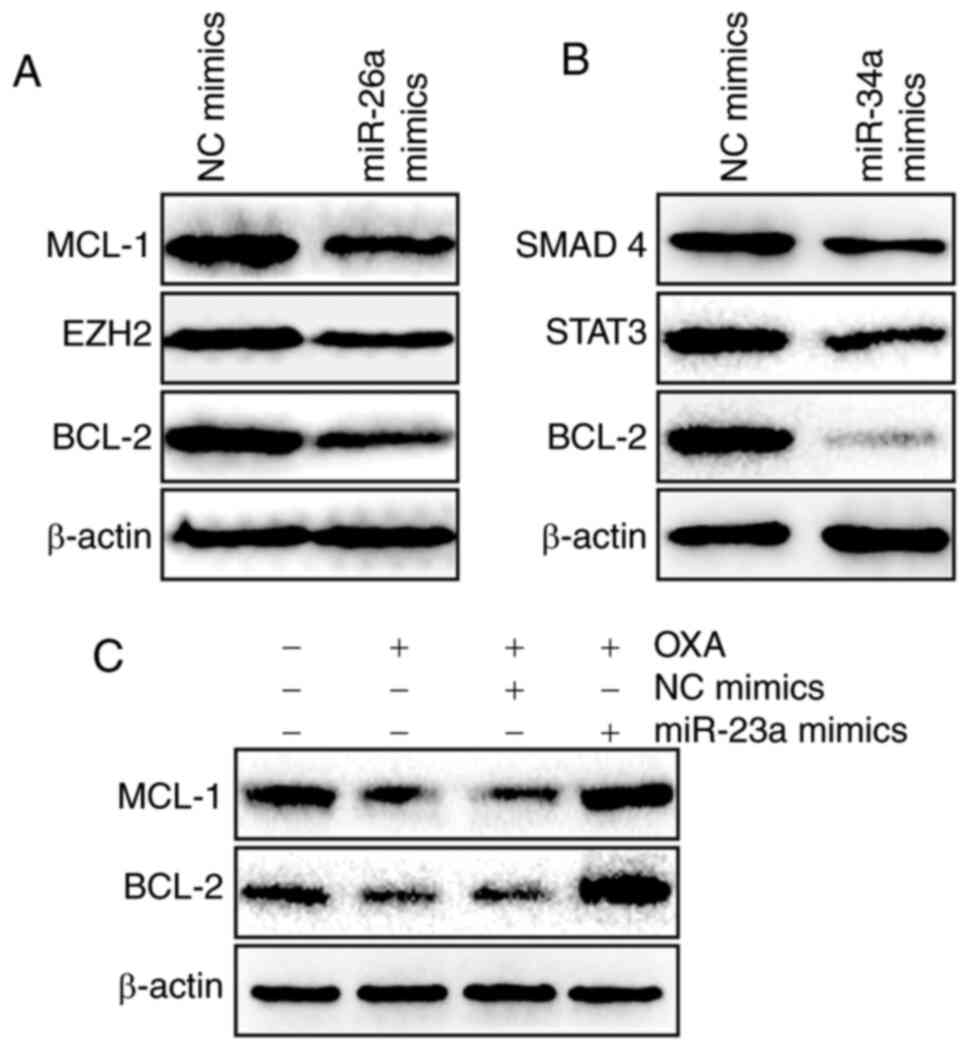
The possible molecular mechanism of how miR-23a, 26a
and 34a modulate OXA sensitivity was further investigated.
According to previous studies, miR-26a and 34a may activate BCL-2,
MCL-1, EZH2, SMAD 4 and STAT3 (54–58).
In the present study, it was also demonstrated that miR-26a could
decrease the expression levels of MCL-1, EZH2 and BCL-2 (Fig. 8A) and miR-34a could decrease the
expression levels of SMAD4, STAT3 and BCL-2 (Fig. 8B). However, high level of miR-23a
could reverse the OXA-induced suppression of MCL-1 and BCL-2
(Fig. 8C). These data possibly
explain why P53WT-induced miRs promote OXA sensitivity
under normoxic conditions but hypoxia enhances OXA resistance in
CRC cells.
Since OXA is a major antitumor drug for CRC
chemotherapy, finding useful biomarkers and potential molecular
mechanisms of OXA resistance is significant for adjusting the
treatment regimen for patients with CRC. The hypoxic tumor
microenvironment has a pivotal influence on behavior of tumor
cells. Impaired drug penetration into hypoxic regions of tumors and
adaptive cellular response to hypoxia are considered to account for
the reduction of cytotoxicity of numerous drugs in multiple cancer
types (59–61).
In the present study, the OXA sensitivity of HCT116
and HT29 cells was firstly examined. The cell proliferation
inhibition results clearly showed that hypoxia affects the OXA
sensitivity of both cell lines. Notably, hypoxia caused a greater
impact on HCT116 cells than on HT29 cells. Since the major
difference between these two cell lines is the TP53 genotype
(HCT116 is TP53WT and HT29 is TP53MT), it was
hypothesized that P53 may play a key role in hypoxia-induced OXA
resistance. To test this hypothesis, the P53 expression level was
determined after HCT116 and HT29 cells were treated with 20 µM OXA
or hypoxia and it was found that the P53WT level in
HCT116 cells was significantly increased, while the
P53MT level in HT29 cells did not change considerably.
P53 was then knocked down and drug sensitivity was examined.
Surprisingly, it was found that P53 had dual effects on regulating
the OXA sensitivity of HCT116 cells: It promoted OXA sensitivity
under normoxic conditions and reduced OXA sensitivity under hypoxic
conditions.
miRs are critical transcriptional mediators and
epigenetic regulators in multiple biological activities, including
tumorigenesis, angiogenesis, cell senescence, metabolism and drug
resistance (62–66). There is a great number of miRs in
cells and thousands of human genes are miR targets. Compared with
transcriptional regulation, miR regulation is fast and flexible,
and miRs may regulate cellular behaviors without affecting basic
biological activities. Previous studies have indicated that there
are numerous P53-dependent miRs (67,68).
Most of these miRs have P53 response elements in their promoter
region, such as miR-145, −34s, −202, −1204, −1206, −10b and −23b
(69–72). This suggests that although P53 is
upregulated by OXA and hypoxia, it may activate different miR
groups under these two stimuli, which can explain the distinct
effects of P53 on OXA sensitivity under normoxic and hypoxic
conditions. By referring to previous research findings, 31 OXA- or
hypoxia-induced miRs were selected. Since numerous of these miRs
are P53 dependent (Table I)
(73–76), they are probably involved in the
regulation of resistance by P53. RT-qPCR results indicated that
among these miRs, 6 miRs were induced by OXA, and 8miRs were
induced by hypoxia. The following P53 deprivation experiments
indicated that miR-26a-5p, miR-34a-5p and miR-133a-3p levels were
associated with P53 in OXA treatment conditions, while miR-23a-3p,
miR-107 and miR-205-5p were altered in hypoxia treatment
conditions. Further miR overexpression experiments indicated that
among the aforementioned six miRs, only miR-23a-3p (decreases OXA
resistance), miR-26a-5p (increases OXA sensitivity) and miR-34a-5p
(increases OXA sensitivity) were involved in the OXA response.
These three miRs were then introduced into TP53MT HT29
cells, and a similar phenomenon was observed. P53WT
restoration experiments in HT29 cells clearly indicated that
miR-26a-5p, miR-34a-5p and miR-23a-3p are P53 dependent. Taking
these data together, it could be concluded that in P53WT
HCT116 cells, P53WT protein plays two roles in
regulating cell sensitivity to OXA: Under normoxic conditions, OXA
stimulates P53WT expression and therefore induces
miR-26a-5p and miR-34a-5p, which renders cells sensitive to OXA; by
contrast, under hypoxic condition, although P53WT is
also upregulated, it induces miR-23a and decreases cell sensitivity
to OXA.
Hypoxia activates HIF signaling pathways in cancer
cells, which can transactivate a wide variety of transcripts
including miR transcripts (77). To
further confirm that miR-26a-5p and miR-34a-3p are specifically
induced by hypoxia, echinomycin was used to inactivate HIF-1α, and
it was found that miR-26a-5p and miR-34a-3p could not be sustained
without HIF-1α in both HCT116 and HT29 cells. These data indicated
that although P53WT can be used to regulate these two
miRs under hypoxic conditions, HIF-1α is necessary for their stable
expression.
miRs regulate drug sensitivity through modulation of
numerous cellular apoptosis- and gene transcription-related
factors. For example, Zhou et al (78) reported that miR-26a inhibits bladder
cancer cell proliferation through inhibition of EZH2; Li et
al (79) showed that knocking
down EZH2 promotes OXA-induced cell cytotoxicity in OXA-resistant
HT29 cells; Gao et al (55)
reported that miR-26a inhibits breast cancer cell proliferation
through repression of MCL-1; Yang et al (56) reported that miR-26a decreases Bcl-2
expression and that suppression of miR-26a causes cisplatin
resistance in human non-small cell lung cancer. These findings
supported our statement that P53WT-induced miR-26a
overexpression promotes OXA sensitivity in HCT116 cells. Similarly,
miR-34a may promote OXA sensitivity through interactions with
SMAD4, STAT3 and BCL-2 (58,80–82).
The underlying molecular mechanisms for the association between
P53WT-induced miR-23a overexpression and
hypoxia-mediated acquired OXA resistance were also investigated.
Western blot results clearly showed that OXA suppressed the
expression of the tumor antiapoptotic molecules MCL-1 and BCL-2.
However, administration of miR-23a mimics restored their levels.
Numerous studies have reported the oncogenic and promoting drug
resistance property of miR-23a in cancers (83–86).
Jin et al (39) reported
that hypoxia led to an upregulation of miR-23a in CRC cells. Xu
et al (87) found that
sinomenine exerts an antitumor effect by downregulating miR-23a,
and transfection of miR-23a-3p increased the level of BCL-2 in PC3
cells. Zhang et al (88)
also revealed that miR-23a-3p inhibited the expression of BAX,
promoted the expression of BCL-2 and inhibited the apoptosis of
U937 cells. These investigations were in line with the present
findings.
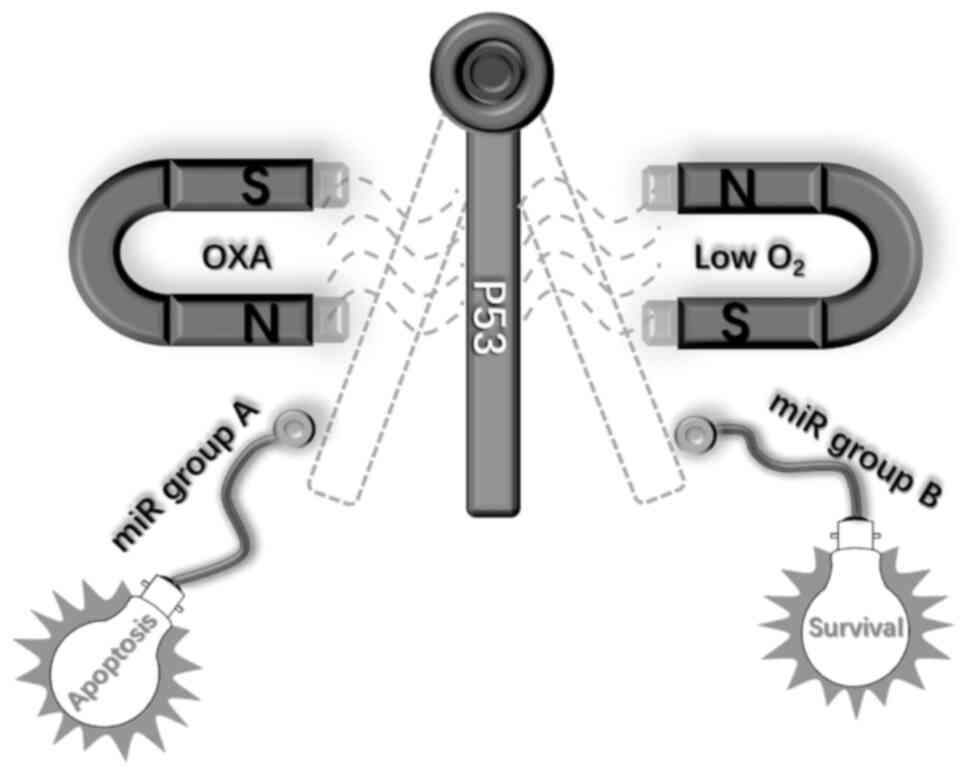
In summary, in the present study, the role of P53 in
OXA-induced cellular apoptosis in CRC was investigated and it was
identified that under normoxic conditions, P53 may promote cell
apoptosis through activation of miR-26a and miR-34a, whereas under
hypoxic conditions, P53 may induce OXA resistance through the
activation of miR-23a. The present findings revealed the dual
function of P53 in regulating cell apoptosis and highlighted the
role of P53-miR interactions in the response of CRC cells to OXA
under normoxic and hypoxic conditions (Fig. 9). These findings may provide deep
insight into the molecular mechanism of antitumor drug resistance
and a novel idea to overcome drug resistance in clinical cancer
treatment. However, to deeply understand the crosstalk between P53
and the miR group, the identification of more research targets,
particularly through data mining and bioinformatics analysis, is
still needed.
Not applicable.
The present study was supported by grants from the National
Natural Science Foundation of China (grant no. 81903380) and the
Wanzhong Biological Technology Ltd. Co (grant no.
3R2210279430).
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
TZ and DS contributed to the conception and design
of the study. JZ and CL contributed to the acquisition, analysis
and interpretation of data. LS analyzed the data and was involved
in performing the experiments. DS drafted the work. TZ revised it
critically for important intellectual content. All authors confirm
the authenticity of all the raw data and read and approved the
final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Cai J and Wang L: Looking back
2018-focused on colorectal cancer. Zhonghua Wei Chang Wai Ke Za
Zhi. 22:9–16. 2019.(In Chinese). PubMed/NCBI
|
|
2
|
Quirke P, Durdey P, Dixon MF and Williams
NS: Local recurrence of rectal adenocarcinoma due to inadequate
surgical resection. Histopathological study of lateral tumour
spread and surgical excision. Lancet. 2:996–999. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hsu HH, Chen MC, Baskaran R, Lin YM, Day
CH, Lin YJ, Tu CC, Vijaya Padma V, Kuo WW and Huang CY: Oxaliplatin
resistance in colorectal cancer cells is mediated via activation of
ABCG2 to alleviate ER stress induced apoptosis. J Cell Physiol.
233:5458–5467. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang C, Zhou Q, Li M, Tong X, Sun J, Qing
Y, Sun L, Yang X, Hu X, Jiang J, et al: Upregulation of CYP2S1 by
oxaliplatin is associated with p53 status in colorectal cancer cell
lines. Sci Rep. 6:330782016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meads MB, Gatenby RA and Dalton WS:
Environment-mediated drug resistance: A major contributor to
minimal residual disease. Nat Rev Cancer. 9:665–674. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martinez-Balibrea E, Martínez-Cardús A,
Ginés A, Ruiz de Porras V, Moutinho C, Layos L, Manzano JL, Bugés
C, Bystrup S, Esteller M and Abad A: Tumor-related molecular
mechanisms of oxaliplatin resistance. Mol Cancer Ther.
14:1767–1776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Plasencia C, Martinez-Balibrea E,
Martinez-Cardús A, Quinn DI, Abad A and Neamati N: Expression
analysis of genes involved in oxaliplatin response and development
of oxaliplatin-resistant HT29 colon cancer cells. Int J Oncol.
29:225–235. 2006.PubMed/NCBI
|
|
8
|
Pedraz-Cuesta E, Christensen S, Jensen AA,
Jensen NF, Bunch L, Romer MU, Brünner N, Stenvang J and Pedersen
SF: The glutamate transport inhibitor DL-Threo-β-Benzyloxyaspartic
acid (DL-TBOA) differentially affects SN38- and oxaliplatin-induced
death of drug-resistant colorectal cancer cells. BMC Cancer.
15:4112015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liao X, Song G, Xu Z, Bu Y, Chang F, Jia
F, Xiao X, Ren X, Zhang M and Jia Q: Oxaliplatin resistance is
enhanced by saracatinib via upregulation Wnt-ABCG1 signaling in
hepatocellular carcinoma. BMC Cancer. 20:312020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hubbi ME and Semenza GL: Regulation of
cell proliferation by hypoxia-inducible factors. Am J Physiol Cell
Physiol. 309:C775–C782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pugh CW and Ratcliffe PJ: Regulation of
angiogenesis by hypoxia: Role of the HIF system. Nat Med.
9:677–684. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gilkes DM, Semenza GL and Wirtz D: Hypoxia
and the extracellular matrix: Drivers of tumour metastasis. Nat Rev
Cancer. 14:430–439. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang YA, Chen YF, Bao Y, Mahara S, Yatim
SMJM, Oguz G, Lee PL, Feng M, Cai Y, Tan EY, et al: Hypoxic tumor
microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote
chemoresistance in colorectal cancer. Proc Natl Acad Sci USA.
115:E5990–E5999. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roberts DL, Williams KJ, Cowen RL,
Barathova M, Eustace AJ, Brittain-Dissont S, Tilby MJ, Pearson DG,
Ottley CJ, Stratford IJ and Dive C: Contribution of HIF-1 and drug
penetrance to oxaliplatin resistance in hypoxic colorectal cancer
cells. Br J Cancer. 101:1290–1297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gariboldi MB, Taiana E, Bonzi MC,
Craparotta I, Giovannardi S, Mancini M and Monti E: The BH3-mimetic
obatoclax reduces HIF-1α levels and HIF-1 transcriptional activity
and sensitizes hypoxic colon adenocarcinoma cells to
5-fluorouracil. Cancer Lett. 364:156–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nijhuis A, Thompson H, Adam J, Parker A,
Gammon L, Lewis A, Bundy JG, Soga T, Jalaly A, Propper D, et al:
Remodelling of microRNAs in colorectal cancer by hypoxia alters
metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet.
26:1552–1564. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu K, Zhan Y, Yuan Z, Qiu Y, Wang H, Fan
G, Wang J, Li W, Cao Y, Shen X, et al: Hypoxia induces drug
resistance in colorectal cancer through the HIF-1α/miR-338-5p/IL-6
feedback loop. Mol Ther. 27:1810–1824. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Therachiyil L, Haroon J, Sahir F, Siveen
KS, Uddin S, Kulinski M, Buddenkotte J, Steinhoff M and
Krishnankutty R: Dysregulated phosphorylation of p53, autophagy and
stemness attributes the mutant p53 harboring colon cancer cells
impaired sensitivity to oxaliplatin. Front Oncol. 10:17442020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shang L and Wei M: Inhibition of SMYD2
sensitized cisplatin to resistant cells in NSCLC through activating
p53 pathway. Front Oncol. 9:3062019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takayama T, Miyanishi K, Hayashi T, Sato Y
and Niitsu Y: Colorectal cancer: Genetics of development and
metastasis. J Gastroenterol. 41:185–192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen
H, Xia D, Xu E, Lai M, Wu Y and Zhang H: Mutations of key driver
genes in colorectal cancer progression and metastasis. Cancer
Metastasis Rev. 37:173–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sui X, Kong N, Wang X, Fang Y, Hu X, Xu Y,
Chen W, Wang K, Li D, Jin W, et al: JNK confers 5-fluorouracil
resistance in p53-deficient and mutant p53-expressing colon cancer
cells by inducing survival autophagy. Sci Rep. 4:46942014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ikediobi ON, Davies H, Bignell G, Edkins
S, Stevens C, O'Meara S, Santarius T, Avis T, Barthorpe S,
Brackenbury L, et al: Mutation analysis of 24 known cancer genes in
the NCI-60 cell line set. Mol Cancer Ther. 5:2606–2612. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park JY, Park SJ, Shim KY, Lee KJ, Kim YB,
Kim YH and Kim SK: Echinomycin and a novel analogue induce
apoptosis of HT-29 cells via the activation of MAP kinases pathway.
Pharmacol Res. 50:201–207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim JS, Lee C, Bonifant CL, Ressom H and
Waldman T: Activation of p53-dependent growth suppression in human
cells by mutations in PTEN or PIK3CA. Mol Cell Biol. 27:662–677.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng X, Sun W, Yu J, Zhou Y, Gu Y, Han J,
Zhou L, Jiang X and Wang C: LINC00460-miR-149-5p/miR-150-5p-mutant
p53 feedback loop promotes oxaliplatin resistance in colorectal
cancer. Mol Ther Nucleic Acids. 22:1004–1015. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maqbool R, Lone SN and Ul Hussain M:
Post-transcriptional regulation of the tumor suppressor p53 by a
novel miR-27a, with implications during hypoxia and tumorigenesis.
Biochem J. 473:3597–3610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Rokavec M, Jiang L, Horst D and
Hermeking H: Antagonistic effects of p53 and HIF1A on microRNA-34a
regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to
mesenchymal transition in colorectal cancer cells.
Gastroenterology. 153:505–520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kiyonari S, Iimori M, Matsuoka K, Watanabe
S, Morikawa-Ichinose T, Miura D, Niimi S, Saeki H, Tokunaga E, Oki
E, et al: The 1,2-diaminocyclohexane carrier ligand in oxaliplatin
induces p53-dependent transcriptional repression of factors
involved in thymidylate biosynthesis. Mol Cancer Ther.
14:2332–2342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moradi Marjaneh R, Khazaei M, Ferns GA,
Avan A and Aghaee-Bakhtiari SH: MicroRNAs as potential therapeutic
targets to predict responses to oxaliplatin in colorectal cancer:
From basic evidence to therapeutic implication. IUBMB Life.
71:1428–1441. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Islam SU, Ahmed MB, Sonn JK, Jin EJ and
Lee YS: PRP4 induces epithelial-mesenchymal transition and drug
resistance in colon cancer cells via activation of p53. Int J Mol
Sci. 23:30922022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Du Y, Wei N, Ma R, Jiang S and Song D: A
miR-210-3p regulon that controls the Warburg effect by modulating
HIF-1α and p53 activity in triple-negative breast cancer. Cell
Death Dis. 11:7312020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nersisyan S, Galatenko A, Chekova M and
Tonevitsky A: Hypoxia-induced miR-148a downregulation contributes
to poor survival in colorectal cancer. Front Genet. 12:6624682021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ullmann P, Qureshi-Baig K, Rodriguez F,
Ginolhac A, Nonnenmacher Y, Ternes D, Weiler J, Gäbler K, Bahlawane
C, Hiller K, et al: Hypoxia-responsive miR-210 promotes
self-renewal capacity of colon tumor-initiating cells by repressing
ISCU and by inducing lactate production. Oncotarget. 7:65454–65470.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Evert J, Pathak S, Sun XF and Zhang H: A
study on effect of oxaliplatin in MicroRNA expression in human
colon cancer. J Cancer. 9:2046–2053. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamakuchi M, Lotterman CD, Bao C, Hruban
RH, Karim B, Mendell JT, Huso D and Lowenstein CJ: P53-induced
microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad
Sci USA. 107:6334–6339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Yu M, Zhao K, He M, Ge W, Sun Y,
Wang Y, Sun H and Hu Y: Upregulation of MiR-205 under hypoxia
promotes epithelial-mesenchymal transition by targeting ASPP2. Cell
Death Dis. 7:e25172016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin F, Yang R, Wei Y, Wang D, Zhu Y, Wang
X, Lu Y, Wang Y, Zen K and Li L: HIF-1α-induced miR-23a~27a~24
cluster promotes colorectal cancer progression via reprogramming
metabolism. Cancer Lett. 440–441. 211–222. 2019.
|
|
40
|
Qian X, Yu J, Yin Y, He J, Wang L, Li Q,
Zhang LQ, Li CY, Shi ZM, Xu Q, et al: MicroRNA-143 inhibits tumor
growth and angiogenesis and sensitizes chemosensitivity to
oxaliplatin in colorectal cancers. Cell Cycle. 12:1385–1394. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He C, Wang L, Zhang J and Xu H:
Hypoxia-inducible microRNA-224 promotes the cell growth, migration
and invasion by directly targeting RASSF8 in gastric cancer. Mol
Cancer. 16:352017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen HY, Lin YM, Chung HC, Lang YD, Lin
CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, et al: miR-103/107
promote metastasis of colorectal cancer by targeting the metastasis
suppressors DAPK and KLF4. Cancer Res. 72:3631–3641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun Z, Zhang Q, Yuan W, Li X, Chen C, Guo
Y, Shao B, Dang Q, Zhou Q, Wang Q, et al: MiR-103a-3p promotes
tumour glycolysis in colorectal cancer via hippo/YAP1/HIF1A axis. J
Exp Clin Cancer Res. 39:2502020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tang Y, Weng X, Liu C, Li X and Chen C:
Hypoxia enhances activity and malignant behaviors of colorectal
cancer cells through the STAT3/MicroRNA-19a/PTEN/PI3K/AKT axis.
Anal Cell Pathol (Amst). 2021:41324882021.PubMed/NCBI
|
|
45
|
Kim CW, Oh ET, Kim JM, Park JS, Lee DH,
Lee JS, Kim KK and Park HJ: Hypoxia-induced microRNA-590-5p
promotes colorectal cancer progression by modulating matrix
metalloproteinase activity. Cancer Lett. 416:31–41. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu K, Chen G, Qiu Y, Yuan Z, Li H, Yuan X,
Sun J, Xu J, Liang X and Yin P: miR-503-5p confers drug resistance
by targeting PUMA in colorectal carcinoma. Oncotarget.
8:21719–21732. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Saieva L, Barreca MM, Zichittella C, Prado
MG, Tripodi M, Alessandro R and Conigliaro A: Hypoxia-induced
miR-675-5p supports β-catenin nuclear localization by regulating
GSK3-β activity in colorectal cancer cell lines. Int J Mol Sci.
21:38322020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Costa V, Lo Dico A, Rizzo A, Rajata F,
Tripodi M, Alessandro R and Conigliaro A: MiR-675-5p supports
hypoxia induced epithelial to mesenchymal transition in colon
cancer cells. Oncotarget. 8:24292–24302. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Poel D, Boyd LNC, Beekhof R, Schelfhorst
T, Pham TV, Piersma SR, Knol JC, Jimenez CR, Verheul HMW and
Buffart TE: Proteomic analysis of miR-195 and miR-497 replacement
reveals potential candidates that increase sensitivity to
oxaliplatin in MSI/P53wt colorectal cancer cells. Cells.
8:11112019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dong Y, Zhao J, Wu CW, Zhang L, Liu X,
Kang W, Leung WW, Zhang N, Chan FK, Sung JJ, et al: Tumor
suppressor functions of miR-133a in colorectal cancer. Mol Cancer
Res. 11:1051–1060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Moriondo G, Scioscia G, Soccio P, Tondo P,
De Pace CC, Sabato R, Foschino Barbaro MP and Lacedonia D: Effect
of hypoxia-induced micro-RNAs expression on oncogenesis. Int J Mol
Sci. 23:62942022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang X, Lan Z, He J, Lai Q, Yao X, Li Q,
Liu Y, Lai H, Gu C, Yan Q, et al: LncRNA SNHG6 promotes
chemoresistance through ULK1-induced autophagy by sponging
miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 19:2342019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu J, Ke F, Chen T, Zhou Q, Weng L, Tan
J, Shen W, Li L, Zhou J, Xu C, et al: MicroRNAs that regulate PTEN
as potential biomarkers in colorectal cancer: A systematic review.
J Cancer Res Clin Oncol. 146:809–820. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu M, Chen X, Lin K, Zeng K, Liu X, Xu X,
Pan B, Xu T, Sun L, He B, et al: lncRNA SNHG6 regulates EZH2
expression by sponging miR-26a/b and miR-214 in colorectal cancer.
J Hematol Oncol. 12:32019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gao J, Li L, Wu M, Liu M and Xie X, Guo J,
Tang H and Xie X: MiR-26a inhibits proliferation and migration of
breast cancer through repression of MCL-1. PLoS One. 8:e651382013.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang Y, Zhang P, Zhao Y, Yang J, Jiang G
and Fan J: Decreased MicroRNA-26a expression causes cisplatin
resistance in human non-small cell lung cancer. Cancer Biol Ther.
17:515–525. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Werner TV, Hart M, Nickels R, Kim YJ,
Menger MD, Bohle RM, Keller A, Ludwig N and Meese E: MiR-34a-3p
alters proliferation and apoptosis of meningioma cells in vitro and
is directly targeting SMAD4, FRAT1 and BCL2. Aging (Albany NY).
9:932–954. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Velaei K, Samadi N, Barazvan B and
Soleimani Rad J: Tumor microenvironment-mediated chemoresistance in
breast cancer. Breast. 30:92–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Scholten DJ II, Timmer CM, Peacock JD,
Pelle DW, Williams BO and Steensma MR: Down regulation of Wnt
signaling mitigates hypoxia-induced chemoresistance in human
osteosarcoma cells. PLoS One. 9:e1114312014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Muz B, Kusdono HD, Azab F, de la Puente P,
Federico C, Fiala M, Vij R, Salama NN and Azab AK: Tariquidar
sensitizes multiple myeloma cells to proteasome inhibitors via
reduction of hypoxia-induced P-gp-mediated drug resistance. Leuk
Lymphoma. 58:2916–2925. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mirnezami AH, Pickard K, Zhang L, Primrose
JN and Packham G: MicroRNAs: Key players in carcinogenesis and
novel therapeutic targets. Eur J Surg Oncol. 35:339–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Munk R, Panda AC, Grammatikakis I, Gorospe
M and Abdelmohsen K: Senescence-associated MicroRNAs. Int Rev Cell
Mol Biol. 334:177–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vienberg S, Geiger J, Madsen S and
Dalgaard LT: MicroRNAs in metabolism. Acta Physiol (Oxf).
219:346–361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11:252019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tiwari A, Mukherjee B and Dixit M:
MicroRNA key to angiogenesis regulation: MiRNA biology and therapy.
Curr Cancer Drug Targets. 18:266–277. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Oikawa T, Otsuka Y and Sabe H:
p53-dependent and -independent epithelial integrity: beyond miRNAs
and metabolic fluctuations. Cancers (Basel). 10:1622018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Blume CJ, Hotz-Wagenblatt A, Hüllein J,
Sellner L, Jethwa A, Stolz T, Slabicki M, Lee K, Sharathchandra A,
Benner A, et al: p53-dependent non-coding RNA networks in chronic
lymphocytic leukemia. Leukemia. 29:2015–2023. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ueno T, Toyooka S, Fukazawa T, Kubo T, Soh
J, Asano H, Muraoka T, Tanaka N, Maki Y, Shien K, et al:
Preclinical evaluation of microRNA-34b/c delivery for malignant
pleural mesothelioma. Acta Med Okayama. 68:23–26. 2014.PubMed/NCBI
|
|
70
|
Suh SO, Chen Y, Zaman MS, Hirata H,
Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, et
al: MicroRNA-145 is regulated by DNA methylation and p53 gene
mutation in prostate cancer. Carcinogenesis. 32:772–778. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Huang Y, Kesselman D, Kizub D,
Guerrero-Preston R and Ratovitski EA: Phospho-ΔNp63α/microRNA
feedback regulation in squamous carcinoma cells upon cisplatin
exposure. Cell Cycle. 12:684–697. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bisio A, De Sanctis V, Del Vescovo V,
Denti MA, Jegga AG, Inga A and Ciribilli Y: Identification of new
p53 target microRNAs by bioinformatics and functional analysis. BMC
Cancer. 13:5522013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto
T, Kato S and Miyazono K: Modulation of microRNA processing by p53.
Nature. 460:529–533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kulshreshtha R, Ferracin M, Wojcik SE,
Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM,
Negrini M, et al: A microRNA signature of hypoxia. Mol Cell Biol.
27:1859–1867. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liao JM, Cao B, Zhou X and Lu H: New
insights into p53 functions through its target microRNAs. J Mol
Cell Biol. 6:206–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Rokavec M, Li H, Jiang L and Hermeking H:
The p53/microRNA connection in gastrointestinal cancer. Clin Exp
Gastroenterol. 7:395–413. 2014.PubMed/NCBI
|
|
77
|
Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L,
Wang H, Huang C and Sun S: Hypoxia-induced down-regulation of
microRNA-34a promotes EMT by targeting the Notch signaling pathway
in tubular epithelial cells. PLoS One. 7:e307712012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhou B, Wei E, Shi H, Huang J, Gao L,
Zhang T, Wei Y and Ge B: MiR-26a inhibits cell proliferation and
induces apoptosis in human bladder cancer through regulating EZH2
bioactivity. Int J Clin Exp Pathol. 10:11234–11241. 2017.PubMed/NCBI
|
|
79
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemoresistance through EZH2. Mol Cancer Ther. 16:739–751.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC,
Yao L and Qiao PF: miR-34a mediates oxaliplatin resistance of
colorectal cancer cells by inhibiting macroautophagy via
transforming growth factor-β/Smad4 pathway. World J Gastroenterol.
23:1816–1827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shi X, Kaller M, Rokavec M, Kirchner T,
Horst D and Hermeking H: Characterization of a
p53/miR-34a/CSF1R/STAT3 feedback loop in colorectal cancer. Cell
Mol Gastroenterol Hepatol. 10:391–418. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wu T, Wang Z, Liu Y, Mei Z, Wang G, Liang
Z, Cui A, Hu X, Cui L, Yang Y and Liu CY: Interleukin 22 protects
colorectal cancer cells from chemotherapy by activating the STAT3
pathway and inducing autocrine expression of interleukin 8. Clin
Immunol. 154:116–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Jahid S, Sun J, Edwards RA, Dizon D,
Panarelli NC, Milsom JW, Sikandar SS, Gümüs ZH and Lipkin SM:
miR-23a promotes the transition from indolent to invasive
colorectal cancer. Cancer Discov. 2:540–553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Huang S, He X, Ding J, Liang L, Zhao Y,
Zhang Z, Yao X, Pan Z, Zhang P, Li J, et al: Upregulation of
miR-23a approximately 27a approximately 24 decreases transforming
growth factor-beta-induced tumor-suppressive activities in human
hepatocellular carcinoma cells. Int J Cancer. 123:972–978. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li X, Li X, Liao D, Wang X, Wu Z, Nie J,
Bai M, Fu X, Mei Q and Han W: Elevated microRNA-23a expression
enhances the chemoresistance of colorectal cancer cells with
microsatellite instability to 5-fluorouracil by directly targeting
ABCF1. Curr Protein Pept Sci. 16:301–309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Peng F, Zhang H, Du Y and Tan P: miR-23a
promotes cisplatin chemoresistance and protects against
cisplatin-induced apoptosis in tongue squamous cell carcinoma cells
through twist. Oncol Rep. 33:942–950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xu F, Li Q, Wang Z and Cao X: Sinomenine
inhibits proliferation, migration, invasion and promotes apoptosis
of prostate cancer cells by regulation of miR-23a. Biomed
Pharmacother. 112:1085922019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang YS, Wang MY, Zhang WL and Tang CH:
Proliferation, migration and apoptosis of acute myeloid leukemia
cells regulated by mir-23a-3p targeting SMC1A and the mechanism.
Zhonghua Zhong Liu Za Zhi. 41:753–759. 2019.(In Chinese).
PubMed/NCBI
|