Introduction
Uveal melanoma (UM) is the most common intraocular
malignancy among adults. Despite adequate and early primary tumor
treatment, almost 50% of patients ultimately develop metastasis
(1). Patients with metastasis have
a disease-related death within 1 year (4 to 15 months), due to poor
response to any treatments (2).
Various features has been are known to related to UM metastasis,
including larger tumor diameter and thickness, epithelioid cell
type, loss of chromosome 3 heterozygosity, preferentially expressed
antigen in melanoma expression, and loss of BRCA1-associated
protein 1 (BAP1) mutations (3). UMs
can be divided into Class 1 with low metastatic risk and Class 2
with high metastatic risk based on gene expression profiling
including 12-genes expression (4).
However, there is still a subset of patients categorized as Class 1
developing metastasis. It is necessary to identify additional and
reliable biomarkers for metastatic prediction and potential
therapeutic targets in UM.
Stimulator of interferon genes (STING, also known as
TMEM173) is a transmembrane protein located in the endoplasmic
reticulum and mitochondria, and exists in immune-related tissues,
hematological malignancy and solitary tumor (5). An increasing number of studies
demonstrated that in certain tumors, such as skin melanoma, breast
cancer and gastrointestinal cancers, STING expression is lower than
that in normal tissues and is positively associated with patients'
prognosis by promoting intrinsic antitumour immunity (6–8).
However, the function of STING in tumors is complicated and
controversial. In other tumors, STING levels increase and are
negatively related to patient survival (9). STING can play a pro-tumorigenic role
when activated by chemotherapy agents and facilitates cervical
cancer progression during chronic inflammation (10,11).
STING-dependent DNA sensing pathways suppress ferroptosis and
promotes pancreatic tumorigenesis (12). In chromosomally unstable tumor
cells, activated STING promotes cell invasion and metastasis
(13). Increasing evidences
demonstrated that STING is involved in tumorigenesis and
metastasis.
In this context from literature review, it was
observed that there had not been any studies on STING in UM.
Therefore, a preliminary analysis on the relationship between STING
and UM was performed by using publicly available data from The
Cancer Genome Atlas (TCGA). In the present study, it was revealed
that patients with higher STING expression had significantly
shorter overall and disease-free survival times than those with
lower STING expression. In addition, a significant correlation was
identified between high risks of metastasis and higher expression
of STING. This meaningful finding and the emerging controversial
function of STING prompted the authors to investigate further its
possible contribution in UM. Subsequently, this was investigated in
patients with UM and cell lines and it was attempted to unveil the
potential of STING. It was further confirmed that STING is abundant
in UM tissues and cell lines and upregulated in UM tissues compared
with para-UM tissues (choroid tissues). Increasing STING was found
to promote the invasion and migration of UM cells in vitro
by enhancing p38 mitogen-activated protein kinase (p38-MAPK)
signaling. These findings suggested that STING plays a crucial role
in the development and metastasis of UM, and inhibiting the
STING-p38-MAPK pathway in intrinsic UM cells can be a potential
therapeutic target.
Materials and methods
TCGA database analysis
The RNA-seq data and clinical information of 80
patients with UM were obtained from TCGA database (https://portal.gdc.cancer.gov/). Data analysis
was performed using R ×64 4.0.4 software (https://www.rstudios.co/). Overall survival (OS) and
disease-free survival (DFS) analyses of STING expression were
performed using the Gene Expression Profiling Interactive Analysis
database (http://gepia.cancer-pku.cn) (14).
Patient tissues and cell cultures
A total of 20 clinical tissues were collected from
patients who underwent primary enucleation at Eye & ENT
Hospital of Fudan University (Shanghai, China), diagnosed with UM
between January 2013 and December 2019. Among these, five were
metastatic UM tissues obtained from patients who developed
metastasis in the liver, lungs or other distant organs. By
contrast, the other five were non-metastatic UM tissues obtained
from patients who did not develop metastasis after more than five
years of follow-up time. Retrospective data on survival and
metastasis were obtained from medical records updating patients'
follow-up information semi-annually. The remaining ten tissues
comprised five primary UM tissues with surrounding para-UM tissues
(choroid tissues), which were used to compare UM tissues and normal
tissues. Written informed consent was obtained by all patients for
using their tissues and information in researches. The present
study was approved by The Ethics Committee of Eye & ENT
Hospital of Fudan University (approval no. 2020044-1; Shanghai,
China). Tissues were collected by an experienced pathologist
immediately after enucleation and stored in liquid nitrogen. In the
present study, none of the patients had received any treatment
prior to enucleation. Clinicopathological characteristics
(including age and sex distribution) are included in Table II.
 | Table II.Correlations between clinical
characteristics of patients with uveal melanoma and the expression
of STING. |
Table II.
Correlations between clinical
characteristics of patients with uveal melanoma and the expression
of STING.
|
|
| Level of STING
mRNA |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
characteristics | Number | High | Low | χ2 | P-value |
|---|
| Sex |
|
|
| 0.08119 | 0.7757 |
|
Male | 45 | 27 | 18 |
|
|
|
Female | 35 | 19 | 16 |
|
|
| Age, years |
|
|
| 0.05115 | 0.8211 |
|
≤60 | 40 | 24 | 16 |
|
|
|
>60 | 40 | 22 | 18 |
|
|
| Episcleral
invasion |
|
|
| a1.780 | 0.1821 |
|
Yes | 7 | 6 | 1 |
|
|
| No | 68 | 35 | 33 |
|
|
| Histopathological
features |
|
|
| a6.537 | 0.0106 |
|
Epithelioid cell | 13 | 11 | 2 |
|
|
| Spindle
cell | 30 | 11 | 19 |
|
|
| Clinical stage |
|
|
| 8.345 | 0.0039 |
| II | 39 | 17 | 22 |
|
|
| III +
IV | 40 | 28 | 12 |
|
|
| Tumor status |
|
|
| 6.041 | 0.014 |
|
Tumor-free | 61 | 31 | 30 |
|
|
|
Tumor | 18 | 15 | 3 |
|
|
| Ciliary body
location |
|
|
| 2.494 | 0.1143 |
|
Yes | 24 | 17 | 7 |
|
|
| No | 56 | 29 | 27 |
|
|
The UM cell lines (Mel 202, 92.1, Mel270, Omm2.2,
Omm2.3, Omm1 and Omm2.5) used in the present experiments were
kindly provided by WuXi AppTec (https://www.wuxiapptec.com/zh-cn). THP-1 (Human
Monocytic Cell Line), 293T (Human Embryonic Kidney Cell Line) and
ARPE-19 (Adult Retinal Pigment Epithelial Cell Line) were purchased
from the American Type Culture Collection. The UM cell lines, THP-1
and ARPE-19 were maintained in Roswell Park Memorial Institute 1640
(RPMI-1640) medium supplemented with 10% fetal bovine serum (FBS)
and 1% penicillin-streptomycin (PS) and 293T in Dulbecco's Modified
Eagle Medium (DMEM; all from Gibco; Thermo Fisher Scientific, Inc.)
with 10% FBS and 1% PS. All the cell lines were cultured in a
humidified incubator at 37°C under 5% CO2. These
procedures were conducted in accordance with the highest ethical
standards and best laboratory practices to ensure the validity and
reliability of the findings.
Reagents
Primary antibodies targeting STING (cat no. 13647),
p38-MAPK (cat. no. 8690), phosphorylated p38-MAPK (cat. no. 4511)
and GAPDH (cat. no. 2118) were obtained from Cell Signaling
Technology, Inc. The Anti-Rabbit Secondary antibody (cat. no.
DM-001) Detection Module for Wes was purchased from ProteinSimple.
The compound SB203580 (cat. no. S1076) was purchased from Selleck
Chemicals. Wes Separation kits (12–230 kDa; cat. no. SM-W004;
ProteinSimple) were used to perform our protein immunoassay
procedures.
Transfection
Short hairpin RNA (shRNA) sequences (Genomeditech)
were designed to downregulate STING expression, named shSTING, and
the scramble shRNA control was named Scramble. The shRNA sequence
targeting human STING and the scramble shRNA control sequences were
as follows: 5′-TCTCAAGAGAAATCCGTGCGGA-3′ (shSTING), and
5′-TTCTCCGAACGTGTCACGT-3′ (Scramble). The double strands of shRNA
were inserted into lentiviral vector through the pGMLV-SC5-PURO
RNAi packaging plasmid. The concentrations of the two packaged
lentiviral vectors were 5×108 TU/ml. Human full-length
STING cDNA was cloned into the expression plasmid
pHBLV-CMV-MCS-3FLAG-EF1-ZsGreen-T2A-PURO by Hanbio Biotechnology
Co. Ltd. to upregulate STING (STING+), and the empty
pHBLV-PURO lentiviral vector was used as negative control
(Control). The concentrations of the two packaged lentiviral
vectors were 4.5×108 TU/ml and 3×108 TU/ml,
respectively.
UM cells mixed with targeted or negative control
lentiviral vectors were seeded in 24-well-plates
(1.6×105 cells vs. 3.2×106 TU lentiviral
vectors per well), incubated in a humidified incubator at 37°C
under 5% CO2 for 4~6 h until cell attachment, and then
replaced with fresh mediums. Polybrene Reagent (cat. no. GM-040901;
Genomeditech) was used for transfection with the concentration of 5
µg/ml. Stable cells with STING downregulated or upregulated were
selected using puromycin with the concentration of 2 µg/ml. The
transfected cells were used for further experiments after 48 h
incubation.
Wes Separation protein
immunoassay
Proteins were extracted from UM cells or tissues
using RIPA buffer and a phosphatase inhibitor, followed using the
Wes Separation (Wes) system according to the manufacturer's
protocol. The Wes system is a next-generation hand-free capillary
immunoassay platform combining protein separation with sensitive
chemiluminescence and fluorescence immunodetection. This mature
technology has been applied widely in vaccines, biopharmaceutical
purification and other proteins-related researches (15–17).
For immunodetection, a Master Mix solution, sample
buffer, ladder solution, and a chemiluminescence mixture were
prepared. Next, the protein samples were mixed with the sample
buffer and Master Mix and denatured at 95°C for 5 min. The primary
antibodies were diluted in 1:50~200. Ladder solution, protein
samples, primary antibodies and secondary antibodies,
chemiluminescence mixtures, and wash buffer were sequentially added
to the reaction plate on-ice operation. The reaction plate and
capillary tubes were placed into an automated analyzer that
automatically loaded the proteins for separation and
immunodetection in each capillary tube. Finally, immunoreactive
protein bands were visualized according to chemiluminescence
signals and quantified based on the signal intensities, using
Compass Simple Western software (ProteinSimple).
Proliferation experiments
Cell proliferation was assessed using Cell Counting
Kit-8 (CCK-8; Dojindo Laboratories, Inc.) according to the
manufacturer's instructions. A total of 2.5×103 UM cells
were seeded into 96-well plates before transfection and treatment.
10 µl of CCK-8 reagent was added in each well and incubated at 37°C
for 2 h. Cell viability was measured at 450 nm at 0, 24, 48 and 72
h after transfection and drug treatment using microplate reader
(Tecan Group, Ltd.).
Transwell and wound healing
experiments
In brief, hydrated Transwell chambers (pore size,
8-µm, Corning, Inc.) were coated with Matrigel by RPMI-1640 medium
in the cell incubator at 37°C for 2 h. UM cells (1×105)
were seeded into each Transwell chambers in RPMI-1640 medium
without FBS. The Transwell chambers were immersed in 24-well plates
containing RPMI-1640 medium with 10% FBS and the plates were
incubated at 37°C for 24 h. The cells were fixed in chambers using
4% paraformaldehyde at room temperature for 20 min and stained with
1% crystal violet at room temperature for 15 min. The cells
attached to the upper surface of the chambers were wiped out, and
cells that invaded through the pores were counted in six randomly
selected fields (magnification, ×100) using the light microscope
(Leica Microsystems GmbH). The UM cells (5×105) were
placed into six-well plates and cultured for 24 h. A 200-µl pipette
was then used to scrape the cell layer in a straight line, creating
a scratch. The cells were washed twice with RPMI-1640 medium
without FBS, then cultured in RPMI-1640 medium supplemented with 1%
FBS. Images were captured after 48 h, and the cell migration rate
was compared with that of the initial scratch.
In the Transwell assays and wound healing
experiments, although four UM cell lines were treated in different
time and repeated experiments were performed, the backgrounds of
the images compared at the same time/cell lines were consistent
whether in Transwell assays or wound healing experiments.
Statistical analysis
To compare the differences between two groups of
data, the unpaired Student's t-test was employed. MEDCAL software
was used to identify the cutoff value of STING expression through
receiver operating characteristic (ROC) curve analysis. The
Kaplan-Meier test (followed by log-rank test) was used to analyze
OS and DFS. The correlation between the clinical information of
patients with UM and STING expression was assessed using the
chi-square test. All statistical analyses were performed using IBM
SPSS Statistics software (version 25.0; IBM Corp.). *P<0.05 was
considered to indicate a statistically significant
significance.
Results
Upregulation of STING in UM indicates
a poor prognosis
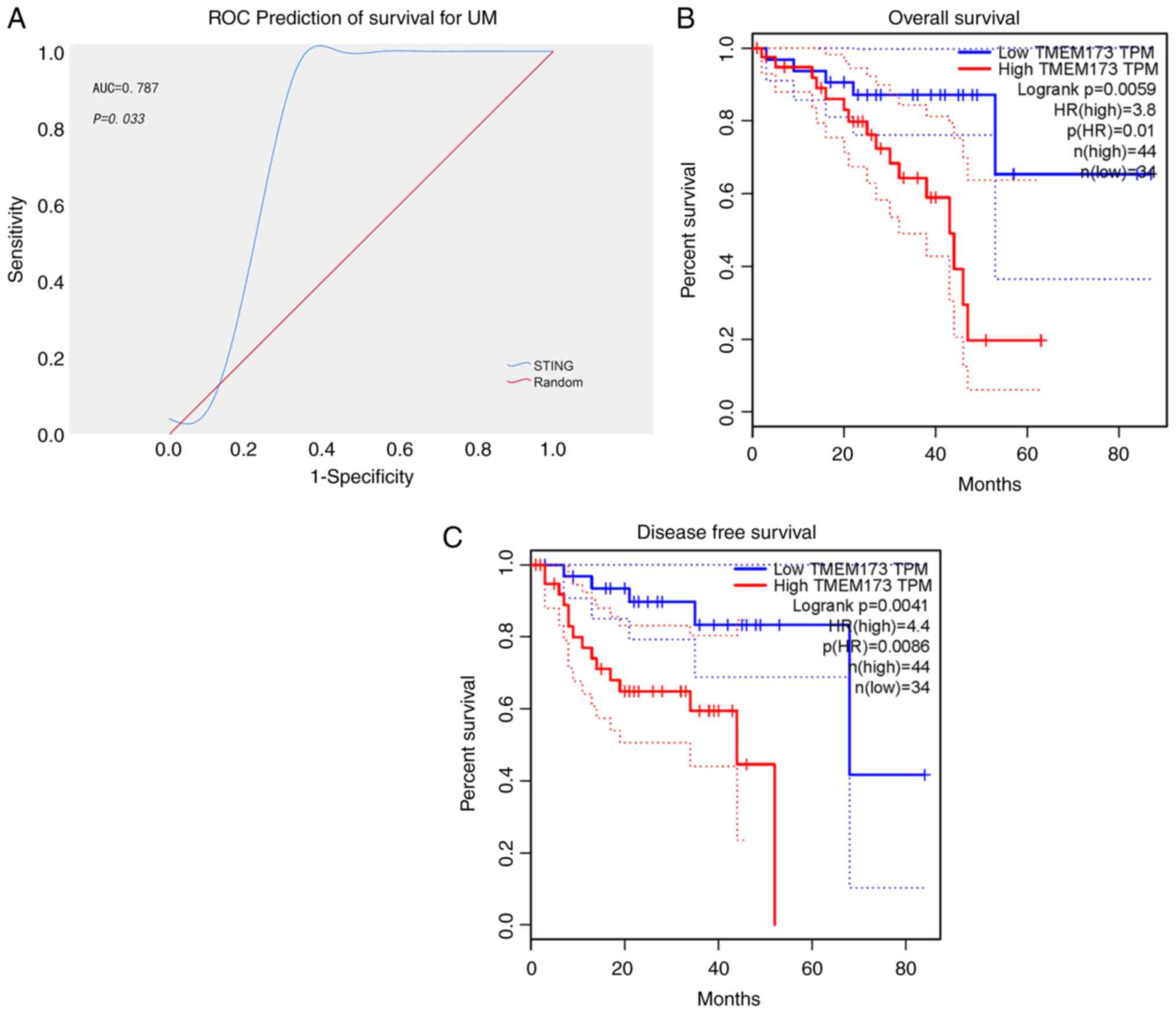
A total of 80 mRNA-seq datasets and the
corresponding clinical information of patients with UM were
retrieved from the TCGA database. An area under the curve was
generated based on STING expression to predict the 5-year survival
of patients. The area under the ROC curve was 0.787, indicating
that expression of STING was a favorable predictor of survival in
patients with UM. The cutoff value for distinguishing between high
and low STING expression was determined to be 10.693 Fragments per
kilobase of transcript per million mapped reads (Fig. 1A and Table I). Survival analysis of patients
with UM demonstrated that those with higher STING expression had
significantly shorter overall and disease-free survival times than
those with lower STING expression (Fig.
1B and C; P<0.01). Furthermore, the chi-square test was
conducted to evaluate the relationship between STING expression and
clinical characteristics. The results revealed a significant
association between high risks of metastasis and higher expression
of STING, including the histological types of epithelioid cells,
higher UM clinical stage, and survival with tumors (Table II; P<0.05). These findings
suggested that upregulation of STING expression indicates a poor
prognosis in patients with UM.
 | Table I.Parameters of receiver operating
characteristic curve based on the expression of stimulator of
interferon genes to predict survival of patients. |
Table I.
Parameters of receiver operating
characteristic curve based on the expression of stimulator of
interferon genes to predict survival of patients.
| Area under the
curve | Standard error | 95% confidence
interval | P (Area=0.5) | Sensitivity | Specificity | Cut-off value
(FPKM) |
|---|
| 0.787 | 0.135 | 0.556–0.933 | 0.033 | 77.78% | 91.67% | ≤10.693 |
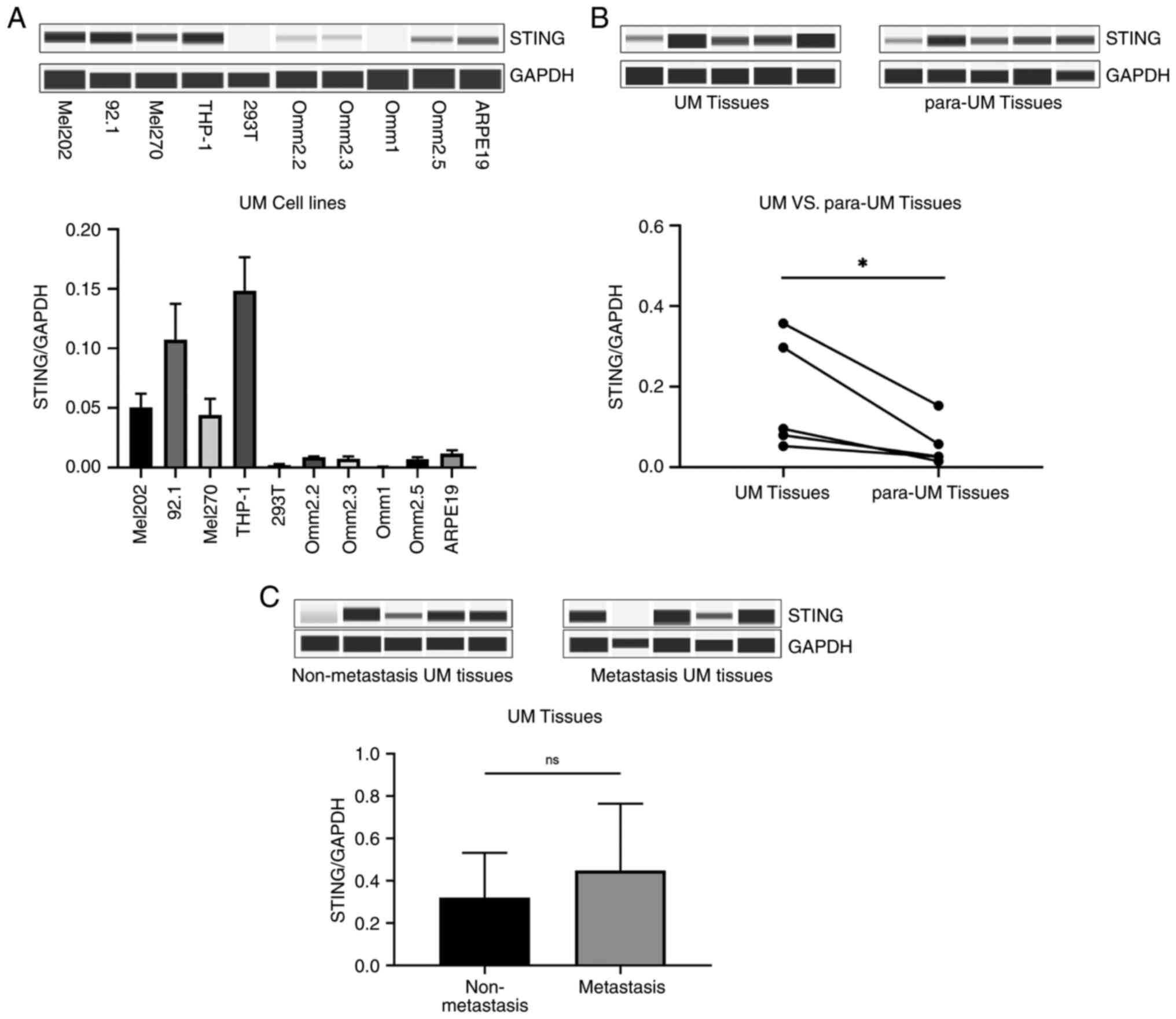
STING is widely expressed in UM and
significantly higher than in normal tissues
To determine the expression levels of STING in UM, a
Wes Separation protein immunoassay was used to detect STING in UM
cell lines, UM tissues and para-UM tissues (choroid tissues). In UM
cell lines, STING exhibited variable expression levels relative to
those in THP-1, 293T and ARPE-19 cells (Fig. 2A). The present findings also
revealed that STING in UM tissues was significantly higher in UM
tissues than in para-UM tissues, suggesting that STING may be
involved in the development of UM (Fig.
2B; P<0.05). However, STING protein expression levels did
not significantly differ between metastatic and non-metastatic UM
tissues (Fig. 2C; P>0.05).
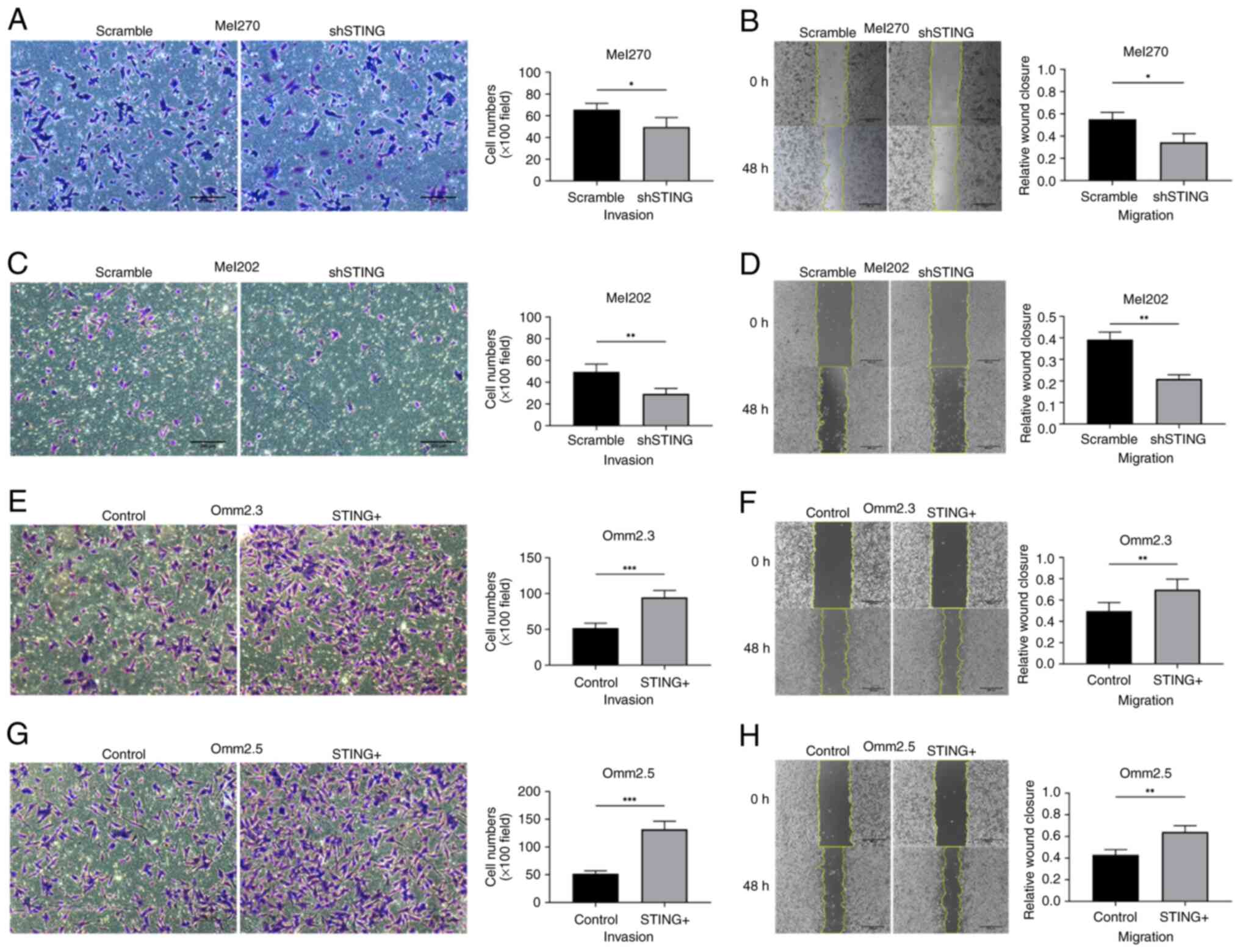
STING promotes invasion and migration
of UM cells
Transwell experiments with Matrigel and wound
healing experiments were performed to evaluate the invasion and
migration ability. Loss-of-function experiments were carried out on
Mel270 and Mel202 cells, which exhibit high STING expression, while
gain-of-function experiments were carried out on Omm2.3 and Omm2.5
cells, with low STING expression levels. First, the shRNA targeting
STING (shSTING) was transfected into Mel270 and Mel202 cells to
downregulate STING with 55~66 and 42~59% reduction respectively,
compared with the Scramble groups (Fig. S1A; P<0.001). Transwell
experiments and wound healing experiments demonstrated that the
downregulation of STING in Mel270 and Mel202 significantly
inhibited their invasive and migratory abilities (Fig. 3A-D; P<0.05). In parallel, the
STING cDNA (STING+) was transfected into Omm2.3 and
Omm2.5 to overexpress STING with 29~32-fold and 15~18-fold
overexpression rate respectively (Fig.
S1B; P<0.001), compared with the Control groups. The
experiment assay revealed that overexpression of STING in Omm2.3
and Omm2.5 significantly promoted their invasive and migratory
abilities (Fig. 3E-H; P<0.05).
However, changes in STING expression did not significantly affect
UM cell proliferation (Fig. S2A-D;
P>0.05).
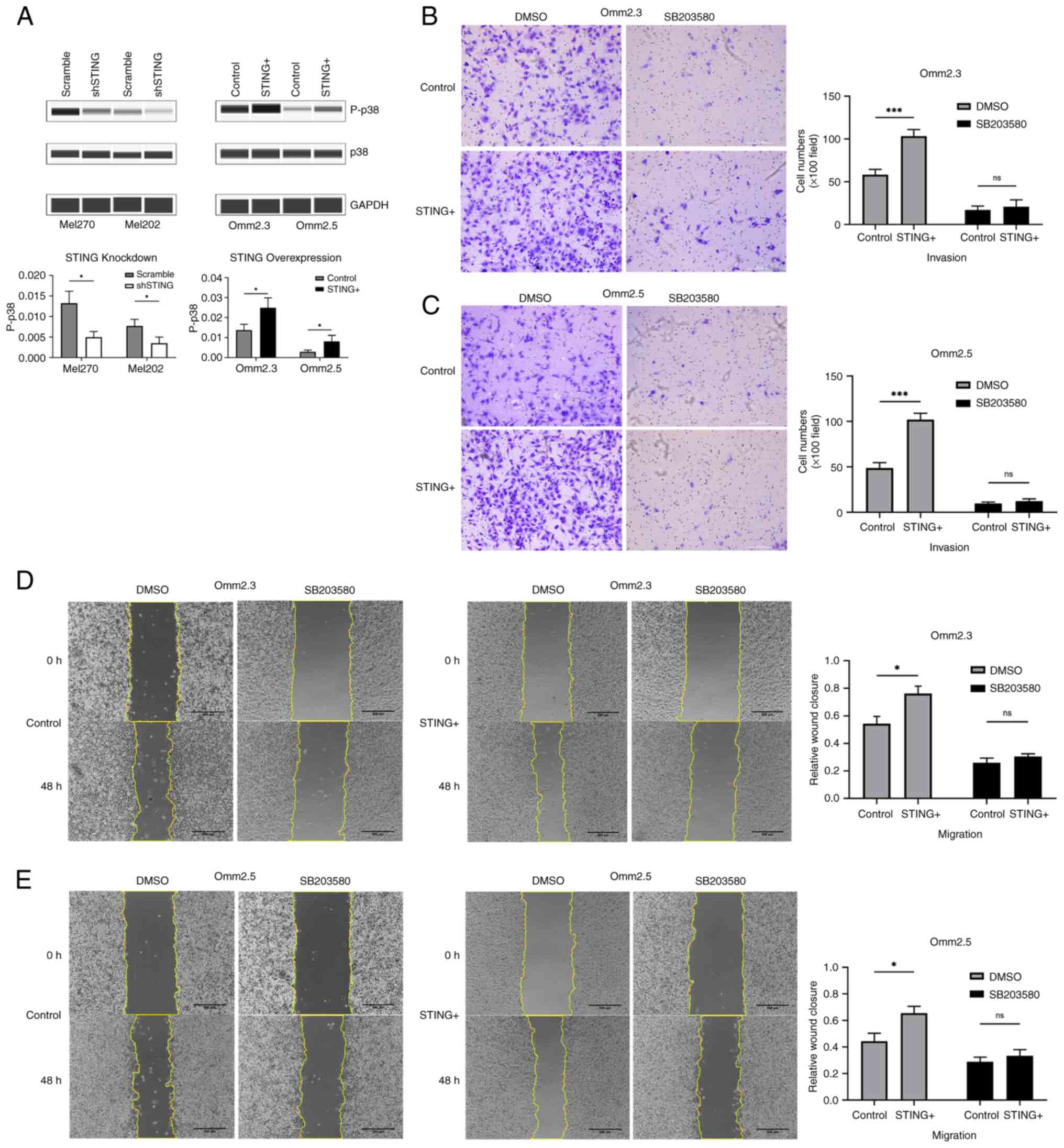
STING promotes the invasion and
migration of UM through p38-MAPK signaling
The roles of MAPKs signaling and BAP1 in UM cell
proliferation and metastasis have been extensively investigated
(18,19). Studies have shown that STING
promotes the development of tumors through canonical and
non-canonical nuclear factor-kappa B (NF-κB) pathways as well as
p21 (13,20,21).
To gain insights into the mechanism of STING-induced UM invasion
and migration, Wes separations were performed to detect the
expression of MAPKs (p38-MAPK, extracellular regulated protein
kinases/ERK, c-Jun N-terminal kinase/JNK), BAP1, NF-κB (p65,
p100/52), and p21 in UM cells. The present results demonstrated
that STING downregulation and overexpression consistently modulated
the levels of phosphorylated p38-MAPK (Fig. 4A), whereas the expression of ERK,
JNK, BAP1, NF-κB (p65, p100/52) and p21 remained unchanged. The
phosphorylation of p38-MAPK decreased when STING was downregulated,
but increased upon STING overexpression. Furthermore, Transwell and
wound healing experiments revealed that SB203580, a potent p38-MAPK
inhibitor (at 10 µM), effectively reversed STING-enhanced cell
invasion and migration (Fig. 4B-E;
P<0.05). Notably, p38-MAPK inhibition had no significant effect
on UM cell proliferation, irrespective of STING expression levels
(Fig. S3A-D; P>0.05).
Discussion
An increasing number of studies have demonstrated
that STING directly influences the tumorigenesis and metastasis of
tumor cells. However, the functions of STING in tumors is
complicated and controversial. Previous studies have consistently
indicated that STING can restrain tumorigenesis and that STING
agonists inhibit tumor development in animal studies and clinical
trials (22,23). However, other studies have also
reported that STING promotes tumorigenesis of certain tumors and
that elevated STING expression is significantly associated with a
poorer prognosis (24,25). On these bases, in the present study,
it was found that upregulation of STING in UM indicates a poor
prognosis according to TCGA data analysis. STING is abundant in
both UM tissue and cell lines and the level of STING was higher in
UM tissues than para-UM tissues (choroid membranes). Moreover,
STING regulated the levels of phosphorylated p38-MAKP and promoted
invasion and migration of UM cells, which could be reversed by a
p38-MAPK inhibitor. All the findings demonstrated that STING
promotes invasion and migration of UM through p38-MAPK
signaling.
Previous studies have revealed that STING expression
in gastrointestinal cancer tissues is markedly lower than that in
normal tissues (7,26). A STING deficiency in tumor cells
accelerates tumor progression, and high STING expression is
associated with an improved prognosis in gastrointestinal cancer,
hepatocellular carcinoma and cervical cancer (27–30).
Activation of cGAS-STING pathway can inhibit the growth of tumor in
numerous animal models by promoting antitumor immunity (8). However, a previous study demonstrated
that STING is higher in squamous carcinoma of the tongue than in
normal tissues and can promote tumor development by increasing
infiltration of regulatory T cells (Tregs) (9). Additionally, high STING expression is
associated with an increased risk of relapse in breast and ovarian
cancers treated with adjuvant chemotherapy (31). Low-grade serous ovarian carcinomas
and serous borderline tumors exhibit uniformly high STING
expression (32). This suggests
that high STING expression reflects pathway activation and that the
mechanisms may vary across different tumors. The present results
are consistent with those of previous studies, wherein the
upregulation of STING in UM indicates a poor prognosis. A possible
explanation for the complicated role of STING is that the
activation patterns and downstream signaling of STING may depend on
the tumor types and cell conditions. STING activation patterns
include phosphorylation, ubiquitination, SUMOylation and
palmitoylation. The downstream signaling includes TANK-binding
kinase 1 (TBK1), canonical/non-canonical NF-κB, DHHC, TRIM56 and so
on (33).
Numerous studies have demonstrated that STING
signaling contributes to cancer development by participating in
immune response, autophagy, cell proliferation and metastasis. For
example, cancer cells migrating to brain activate the STING pathway
and produce inflammatory cytokines in astrocytes to enhance the
growth and chemoresistance of metastatic brain cancers (34). Activated STING in the human
papillomavirus-related carcinogenesis of tongue squamous cells
increases infiltration of Tregs via the c-jun/CCL22 signaling,
potentially leading to tumor immune escape (9). IFI16 promotes cervical cancer
progression in vitro and in vivo through activating
the STING-TBK1-NF-kB pathway (11).
The immune microenvironment is indispensable for STING immune
regulation in tumor. However, UM is an immune-privileged tumor with
an immunosuppressive microenvironment, due to the blood-ocular
barrier and the lack of lymphatic vessels (35). Therefore, the intrinsic effects of
STING on UM cells were investigated in the present study. Cheradame
et al (31) demonstrated
that STING downregulation decreased cell survival and increased
sensitivity to genotoxic treatments in breast cancer cell lines in
a cell-autonomous manner. Bakhoum et al (13) reported that STING, activated by
unstable chromosomes in breast cancer, enhanced cell migration and
invasion, promoting the downstream non-canonical NFκB2 (p52/Rel B)
pathway. Bakhoum et al (36)
also revealed that chromosomal instability is widespread in
high-risk UM and drives UM cell migration in a STING-dependent
manner. The in vitro results of the present study are
consistent with studies that STING promotes invasion and migration
of UM cells, which may be activated by chromosomal instability in
tumor cells.
However, the molecular mechanisms downstream of
STING underlying UM metastasis need to be further elucidated. MAPK
signaling promotes the development of UM cells with Gαq pathway
mutations, and the inhibition of MAPK can suppress progression of
UM effectively (37). The lack of
BAP1 expression in UM tissues significantly predicts metastasis and
indicates a lower metastasis-free survival (19). By contrast, studies have identified
that STING promotes the development of tumors through the canonical
and non-canonical NF-κB pathways and p21 (13,20,21).
Based on the aforementioned studies, it was planned to sift out
potential signaling involved in the downstream pathway of STING to
promote UM, including MAPKs (p38-MAPK, ERK, and JNK), BAP1, NF-κb
(p65 and p100/52) and p21.
The current findings revealed that phosphorylated
p38-MAPK levels decreased in STING-knockdown cells but increased in
STING-overexpressing cells. The p38-MAPK, a member of the canonical
MAPKs family, plays crucial roles in cell proliferation, apoptosis
and motility through phosphorylation and dephosphorylation
processes (38). Previous studies
have demonstrated that p38-MAPK promotes metastasis in various
types of cancers, including hepatocellular carcinoma, head and neck
squamous cell carcinoma, gliomas, renal carcinoma and gastric
adenocarcinoma (39–43). The p38-MAPK signaling upregulates
the expression and secretion of IL-6 in human uveal melanocytes,
and IL-6/STAT3 signaling contributes to UM metastasis via
epithelial-mesenchymal transition (44,45).
Based on previous studies, it was hypothesized that p38-MAPK
signaling is involved in the progression of UM. Functional
experimental results confirmed that STING promotes metastasis of UM
through p38-MAPK signaling. It is worth considering why the
downstream pathways of STING promoting metastasis in UM differ from
breast cancer reported by Bakhoum et al (13). One possible explanation for this is
tumor heterogeneity. Activation patterns and downstream signaling
of STING may depend on the tumor type and cellular conditions.
While the mechanism of STING in UM was being explored by the
authors, other research teams also were paying attention to the
role of STING in UM. Recently, Bakhoum et al (36) reported that the level of STING was
negatively correlated with overall survival and tumor-related
metastasis in UM patients, and treatment with the inhibitor of
STING reduced the migratory phenotype in UM cell line, which is
consistent with the conclusion of the present study. More recently,
on the other hand, Tao et al (46) identified that the photodynamic
polymer couple with cationic platinum agent activated cGAS-STING
pathway improving the survival rate of animal model of UM in
vivo by promoting immune response, which conflicted with the
conclusion of Bakhoum et al (36) and the present conclusions. These
observations suggest that STING plays a pro-tumor role in intrinsic
UM cells but an antitumor role in tumor microenvironment. However,
the UM animal model used in the study by Tao et al (46) was subcutaneous tumor model. It is
widely known that UM is located in the eye, a relative
immune-privileged environment obviously different from other
tissues and organs. This means that the effects of STING on
pro-immunity may not play a decisive role in patients with UM.
Moreover, the bearing tumor in animal experiments in the
aforementioned study originated from OCM-1, a cell line with
controversial origin.
The limitations of the present study include the
absence of in vivo functional experiments for STING and the
small size of the patient cohort used to analyze the correlation
between STING and UM prognosis. Further studies are required to
understand the downstream pathway of p38-MAPK underlying UM
metastasis and the activation pattern of STING in UM. Despite its
preliminary characteristics and limitations, the present study, to
the best of the authors' knowledge, is the first to report the
downstream pathways of STING in UM cells, and to establish that
STING directly promotes the invasion and migration of UM cells via
p38-MAPK signaling in a tumor cell-autonomous manner.
In summary, the results of the present study
indicated that STING promotes the invasion and migration of UM
cells through upregulating the p38-MAPK signaling pathway.
Therefore, STING expression may can serve as a biomarker for
predicting the prognosis of patients with UM, and STING and
p38-MAPK may serve as potential treatment targets for metastatic
UM.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Yu Qi (Zhongshan
Hospital of Fudan University; Shanghai, China) for the help in
analysis of TCGA data.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81970835).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JQ, XZ, JW and BX designed the experiments. XZ, YC
and FM performed the experiments and collected the data. XZ, RM, YC
and BX analyzed and interpreted the data. XZ and FM drafted the
manuscript. BX, RM and FM revised the language of the manuscript.
JQ and JW supervised the present study in ensuring that questions
related to the accuracy or integrity of any part of the work are
appropriately investigated and resolved. XZ, FM and BX confirm the
authenticity of all the raw data. All the authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
2020044-1) by the Institutional Review Board of the Eye & ENT
Hospital of Fudan University (Shanghai, China). Written informed
consent was signed by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
STING
|
stimulator of interferon genes
|
|
UM
|
uveal melanoma
|
|
p38-MAPK
|
p38 mitogen-activated protein
kinase
|
|
TCGA
|
The Cancer Genome Atlas
|
|
CCK-8
|
Cell Counting Kit-8
|
|
BAP1
|
BRCA1-associated protein 1
|
|
TBK-1
|
TANK-binding kinase 1
|
|
RPMI-1640
|
Roswell Park Memorial Institute
1640
|
|
IFN
|
interferon
|
|
FBS
|
fetal bovine serum
|
|
PS
|
penicillin-streptomycin
|
|
shRNA
|
short hairpin RNA
|
|
Wes
|
Wes Separation
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
ROC
|
receiver operating characteristic
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
ERK
|
extracellular regulated protein
kinases
|
|
JNK
|
c-Jun N-terminal kinase
|
References
|
1
|
Kujala E, Mäkitie T and Kivelä T: Very
long-term prognosis of patients with malignant uveal melanoma.
Invest Ophth Vis Sci. 44:4651–4659. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Augsburger JJ, Correa ZM and Shaikh AH:
Effectiveness of treatments for metastatic uveal melanoma. Am J
Ophthalmol. 148:119–127. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smit KN, Jager MJ, de Klein A and Kiliҫ E:
Uveal melanoma: Towards a molecular understanding. Prog Retin Eye
Res. 75:1008002020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Onken MD, Worley LA, Ehlers JP and Harbour
JW: Gene expression profiling in uveal melanoma reveals two
molecular classes and predicts metastatic death. Cancer Res.
64:7205–7209. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang X, Bai XC and Chen ZJ: Structures
and mechanisms in the cGAS-STING innate immunity pathway. Immunity.
53:43–53. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Falahat R, Perez-Villarroel P, Mailloux
AW, Zhu G, Pilon-Thomas S, Barber GN and Mulé JJ: STING signaling
in melanoma cells shapes antigenicity and can promote antitumor
T-cell activity. Cancer Immunol Res. 7:1837–1848. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia T, Konno H, Ahn J and Barber GN:
Deregulation of STING signaling in colorectal carcinoma constrains
DNA damage responses and correlates with tumorigenesis. Cell Rep.
14:282–297. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pantelidou C, Sonzogni O, De Oliveria
Taveira M, Mehta AK, Kothari A, Wang D, Visal T, Li MK, Pinto J,
Castrillon JA, et al: PARP inhibitor efficacy depends on
CD8+ T-cell recruitment via intratumoral STING pathway
activation in BRCA-deficient models of triple-negative breast
cancer. Cancer Discov. 9:722–737. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang D, Xiao-Feng H, Guan-Jun D, Er-Ling
H, Sheng C, Ting-Ting W, Qin-Gang H, Yan-Hong N and Ya-Yi H:
Activated STING enhances Tregs infiltration in the HPV-related
carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal.
Biochim Biophys Acta. 1852:2494–2503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ahn J, Xia T, Konno H, Konno K, Ruiz P and
Barber GN: Inflammation-driven carcinogenesis is mediated through
STING. Nat Commun. 5:51662014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai H, Yan L, Liu N, Xu M and Cai H: IFI16
promotes cervical cancer progression by upregulating PD-L1 in
immunomicroenvironment through STING-TBK1-NF-kB pathway. Biomed
Pharmacother. 123:1097902020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai E, Han L, Liu J, Xie Y, Zeh HJ, Kang
R, Bai L and Tang D: Ferroptotic damage promotes pancreatic
tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway.
Nat Commun. 11:63392020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bakhoum SF, Ngo B, Laughney AM, Cavallo
JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et
al: Chromosomal instability drives metastasis through a cytosolic
DNA response. Nature. 553:467–472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hamm M, Ha S and Rustandi RR: Automated
capillary Western dot blot method for the identity of a 15-valent
pneumococcal conjugate vaccine. Anal Biochem. 478:33–39. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Valdez A and Chen Y: Evaluation of
automated Wes system as an analytical and characterization tool to
support monoclonal antibody drug product development. J Pharm
Biomed Anal. 139:263–268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Borcherding DC, Amin NV, He K, Zhang X,
Lyu Y, Dehner C, Bhatia H, Gothra A, Daud L, Ruminski P, et al: MEK
inhibition synergizes with TYK2 inhibitors in NF1-associated
malignant peripheral nerve sheath tumors. Clin Cancer Res.
29:1592–1604. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Neelature SS and Smalley K: MEK-ing the
most of it: Strategies to Co-target Gαq and MAPK in uveal melanoma.
Clin Cancer Res. 27:1217–1219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szalai E, Wells JR, Ward L and
Grossniklaus HE: Uveal melanoma nuclear BRCA1-associated Protein-1
immunoreactivity is an indicator of metastasis. Ophthalmology.
125:203–209. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Yi S, Zhou J, Zhang Y and Guo F:
The NF-κB subunit RelB regulates the migration and invasion
abilities and the radio-sensitivity of prostate cancer cells. Int J
Oncol. 49:381–392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Belguise K, Kersual N, Kirsch KH,
Mineva ND, Galtier F, Chalbos D and Sonenshein GE: Oestrogen
signalling inhibits invasive phenotype by repressing RelB and its
target BCL2. Nat Cell Biol. 9:470–478. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng L, Liang H, Xu M, Yang X, Burnette B,
Arina A, Li XD, Mauceri H, Beckett M, Darga T, et al:
STING-dependent cytosolic DNA sensing promotes radiation-induced
type I interferon-dependent antitumor immunity in immunogenic
tumors. Immunity. 41:843–852. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding L, Wang Q, Martincuks A, Kearns MJ,
Jiang T, Lin Z, Cheng X, Qian C, Xie S, Kim HJ, et al: STING
agonism overcomes STAT3-mediated immunosuppression and adaptive
resistance to PARP inhibition in ovarian cancer. J Immunother
Cancer. 11:e0056272023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He L, Xiao X, Yang X, Zhang Z, Wu L and
Liu Z: STING signaling in tumorigenesis and cancer therapy: A
friend or foe? Cancer Lett. 402:203–212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Viculin J, Degoricija M, Vilović K, Gabela
I, Franković L, Vrdoljak E and Korac-Prlic J: Elevated tumor
cell-intrinsic STING expression in advanced laryngeal cancer.
Cancers (Basel). 15:35102023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song S, Peng P, Tang Z, Zhao J, Wu W, Li
H, Shao M, Li L, Yang C, Duan F, et al: Decreased expression of
STING predicts poor prognosis in patients with gastric cancer. Sci
Rep. 7:398582017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu C, Guan J, Lu S, Jin Q, Rousseau B, Lu
T, Stephens D, Zhang H, Zhu J, Yang M, et al: DNA sensing in
mismatch Repair-deficient tumor cells is essential for Anti-tumor
immunity. Cancer Cell. 39:96–108. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Parkes EE, Humphries MP, Gilmore E, Sidi
FA, Bingham V, Phyu SM, Craig S, Graham C, Miller J, Griffin D, et
al: The clinical and molecular significance associated with STING
signaling in breast cancer. NPJ Breast Cancer. 7:812021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Zhai Q, Feng X, Chen D, Lu Y, Hu
J, Xie H, Zhou L, Wu J and Zheng S: Cancer cell-intrinsic STING is
associated with CD8 + T-cell infiltration and might serve as a
potential immunotherapeutic target in hepatocellular carcinoma.
Clin Transl Oncol. 23:1314–1324. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kol A, Lubbers JM, Terwindt ALJ, Workel
HH, Plat A, Wisman GBA, Bart J, Nijman HW and De Bruyn M: Combined
STING levels and CD103+ T cell infiltration have significant
prognostic implications for patients with cervical cancer.
Oncoimmunology. 10:19363912021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheradame L, Guerrera IC, Gaston J,
Schmitt A, Jung V, Goudin N, Pouillard M, Radosevic-Robin N,
Modesti M, Judde JG, et al: STING protects breast cancer cells from
intrinsic and genotoxic-induced DNA instability via a
non-canonical, cell-autonomous pathway. Oncogene. 40:6627–6640.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huvila J, Cochrane DR, Ta M, Chow C,
Greening K, Leung S, Karnezis AN, DiFeo A and Huntsman DG: STING
pathway expression in low-grade serous carcinoma of the ovary: An
unexpected therapeutic opportunity? J Pathol Clin Res. 7:548–555.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang H, You Q and Xu X: Targeting
stimulator of interferon genes (STING): A medicinal chemistry
perspective. J Med Chem. 63:3785–3816. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Q, Boire A, Jin X, Valiente M, Er EE,
Lopez-Soto A, Jacob L, Patwa R, Shah H, Xu K, et al:
Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP
transfer. Nature. 533:493–498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niederkorn JY: Immune escape mechanisms of
intraocular tumors. Prog Retin Eye Res. 28:329–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bakhoum MF, Francis JH, Agustinus A,
Earlie EM, Di Bona M, Abramson DH, Duran M, Masilionis I, Molina E,
Shoushtari AN, et al: Loss of polycomb repressive complex 1
activity and chromosomal instability drive uveal melanoma
progression. Nat Commun. 12:54022021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hitchman TD, Bayshtok G, Ceraudo E, Moore
AR, Lee C, Jia R, Wang N, Pachai MR, Shoushtari AN, Francis JH, et
al: Combined inhibition of Gα(q) and MEK enhances therapeutic
efficacy in uveal melanoma. Clin Cancer Res. 27:1476–1490. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hsieh Y, Wu T, Huang C, Hsieh Y, Hwang J
and Liu J: p38 mitogen-activated protein kinase pathway is involved
in protein kinase C alpha-regulated invasion in human
hepatocellular carcinoma cells. Cancer Res. 67:4320–4327. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Junttila MR, Ala-Aho R, Jokilehto T,
Peltonen J, Kallajoki M, Grenman R, Jaakkola P, Westermarck J and
Kähäri V-M: p38alpha and p38delta mitogen-activated protein kinase
isoforms regulate invasion and growth of head and neck squamous
carcinoma cells. Oncogene. 26:5267–5279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Demuth T, Reavie LB, Rennert JL, Nakada M,
Nakada S, Hoelzinger DB, Beaudry CE, Henrichs AN, Anderson EM and
Berens ME: MAP-ing glioma invasion: mitogen-activated protein
kinase kinase 3 and p38 drive glioma invasion and progression and
predict patient survival. Mol Cancer Ther. 6:1212–1222. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li J-K, Chen C, Liu J-Y, Shi J-Z, Liu S-P,
Liu B, Wu D-S, Fang Z-Y, Bao Y, Jiang M-M, et al: Long noncoding
RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma
via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer.
16:1112017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang Q, Lan F, Wang X, Yu Y, Ouyang X,
Zheng F, Han J, Lin Y, Xie Y, Xie F, et al: IL-1β-induced
activation of p38 promotes metastasis in gastric adenocarcinoma via
upregulation of AP-1/c-fos, MMP2 and MMP9. Mol Cancer. 13:182014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu DN, Chen M, Zhang DY, Ye F, McCormick
SA and Chan CC: Interleukin-1beta increases baseline expression and
secretion of interleukin-6 by human uveal melanocytes in vitro via
the p38 MAPK/NF-kappaB pathway. Invest Ophthalmol Vis Sci.
52:3767–3774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gong C, Shen J, Fang Z, et al: Abnormally
expressed JunB transactivated by IL-6/STAT3 signaling promotes
uveal melanoma aggressiveness via epithelial-mesenchymal
transition. Biosci Rep. 38:2018. View Article : Google Scholar
|
|
46
|
Tao H, Tan J, Zhang H, Ren H, Cai Z, Liu
H, Wen B, Du J, Li G, Chen S, et al: cGAS-STING pathway activation
and systemic anti-tumor immunity induction via photodynamic
nanoparticles with potent toxic platinum DNA intercalator against
uveal melanoma. Adv Sci (Weinh). e23028952023. View Article : Google Scholar : PubMed/NCBI
|