Introduction
As the most aggressive primary central nervous
system malignant tumour, glioblastoma (GBM) is a grade IV
astrocytoma comprising 54% of all gliomas, with an incidence of
3.19 per 100,000 individuals in the United States in 2006–2010
(1). Although there are several
therapeutic strategies for GBM treatment, including neurosurgical
therapy, chemotherapy and radiotherapy, the median survival time
for patients with GBM remains at 12–15 months, with a 5-year
survival rate of <5% (1,2).
At present, several molecular biomarkers, including
epidermal growth factor receptor (EGFR), isocitrate dehydrogenase
(IDH), O6-methylguanine DNA methyltransferase (MGMT) and
PTEN have been tested in the clinical setting for patients with GBM
(3). Given that the overall survival
(OS) of patients with GBM remains low, novel molecular biomarkers
and new treatment options are urgently required in order to
determine the developmental mechanisms of GBM.
With the rapid development of high-throughput
sequencing and bioinformatics approaches, the study of oncogene
expression has entered a new stage. To date, an increasing number
of studies have proven that the results of genome-wide tumour
bioinformatic analyses may be used as new biomarkers for diagnosis
and treatment (4–7), which is important for investigating
GBM-associated signaling pathways, such as the MAPK, PI3K and p53
signaling pathways. Multigene signatures have also been confirmed
to predict the prognosis of patients with glioma based on mRNA
expression profiling (8,9).
Alternative splicing (AS) regulates the translation
of mRNA isoforms and gives rise to protein diversity, thus serving
as an important post-transcriptional regulatory mechanism (10). More than 95% of human genes undergo
AS and encode splice variants in transcriptional processes
(11). Increasing evidence indicates
the essential role of AS in the course of oncogenesis, including
tumour cell proliferation, immune escape, angiogenesis and tumour
metastasis (12,13). In addition, specific splicing factor
(SF) genes regulate AS events via binding to pre-mRNAs and yielding
RNA splicing in the tumour microenvironment (14). Abnormal expression of SFs may result
in the activation of oncogenes or the inactivation of cancer
suppressors (15). Therefore, the
role of specific SFs in pathogenesis provides theoretical support
for tumour biological processes, especially at the gene
transcription level (16).
Constructing a prognostic model is essential to elucidate potential
cancer biomarkers (17,18).
Younger patients (median age, 36 years) with GBM
generally have an improved prognosis and commonly carry IDH1
mutations, the cytosine-phosphate-guanine island methylator
phenotype and a gene expression profile of a proneural subgroup.
However, these biomarkers are valid for only a few primary patients
with GBM (19). The present study
constructed a prognostic model with good performance based on AS
events for patients with GBM and plotted SF-AS networks that may
serve as new molecular targets for the prognosis of patients with
GBM.
Materials and methods
Data acquisition
Datasets used in the present study, including RNA
sequencing (RNA-seq) data and corresponding clinical information of
patients with GBM, were downloaded from The Cancer Genome Atlas
(TCGA-GBM; http://tcga-data.nci.nih.gov/tcga/). All subtypes in
this TCGA-GBM dataset, including classical, proneural, mesenchymal
and neural, were analyzed without classification. The expression of
SF genes in the mRNA splicing pathway was obtained from the
SpliceAid2 database (http://www.introni.it/splicing.html). P<0.05 was
considered to indicate a statistically significant difference.
Different splicing types were classified using TCGA
SpliceSeq (20), a Java application,
to investigate the mRNA splicing patterns of RNA-seq and to
identify significant changes in AS events. The percent-splice-in
(PSI) value was calculated using the following formula with
normalized read counts: splice_in(spilce_in+splice_out),
for seven common patterns of AS events, including alternative
acceptor (AA), alternate donor (AD), alternate promoter (AP),
alternate terminator (AT), exon skipping (ES), mutually exclusive
exon (ME) and retained intron (RI) (21). In the current cohort, AS events with
a PSI value >75% were obtained from the TCGA SpliceSeq database.
The PSI value of AS events with standard deviation <1 were
excluded from analysis.
Data analysis, dimension reduction and
model construction
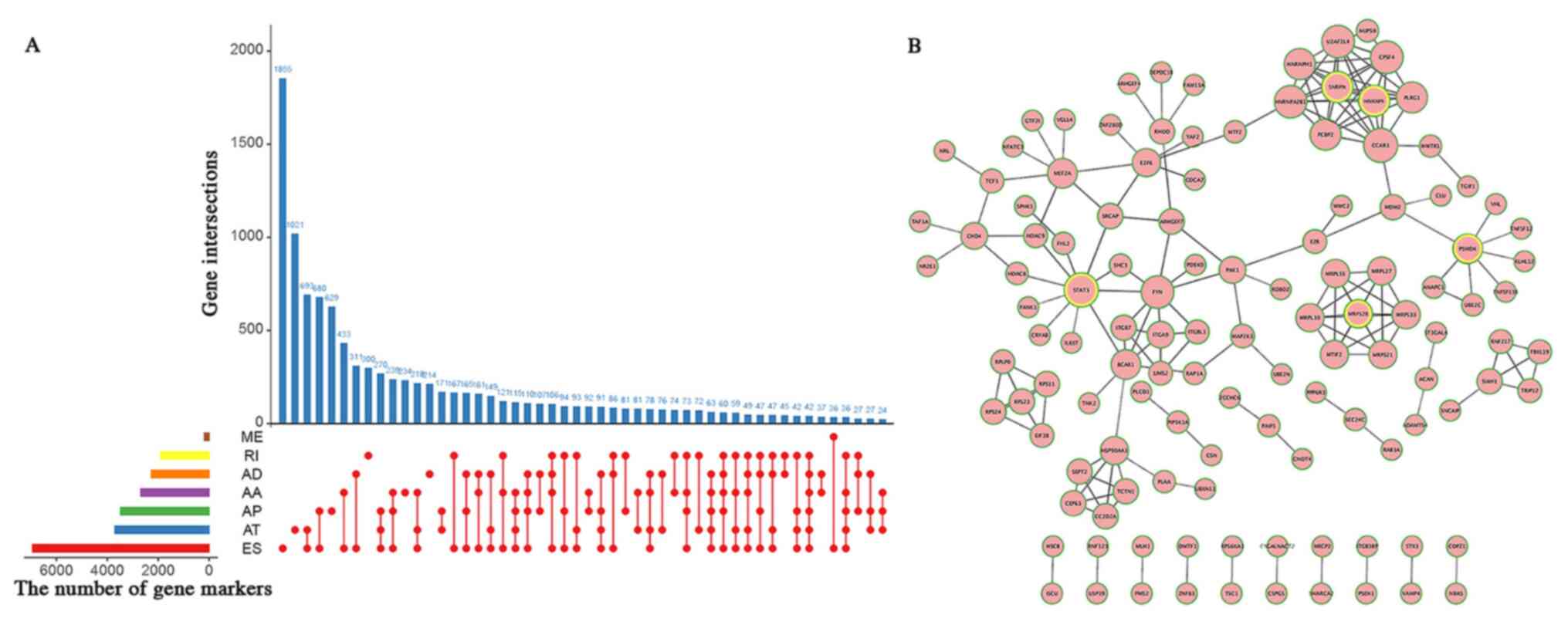
UpSet plot, a novel visualization technique for the
quantitative results of multiple interactive sets (22), was used to visualize various
combinations of the seven aforementioned AS types. To display the
functional interactions of splicing-associated genes, a network was
constructed using the Reactome FI plugin of Cytoscape (version
3.6.1) (23). This application
predicts associations composed of specific genes and integrates
them in a network plot.
Univariate Cox regression was applied to analyze the
association between AS events and OS to disclose the molecular
characteristics of survival-associated AS events. Subsequently, the
top 20 significant AS events of each type were used to develop
prognostic predictor models.
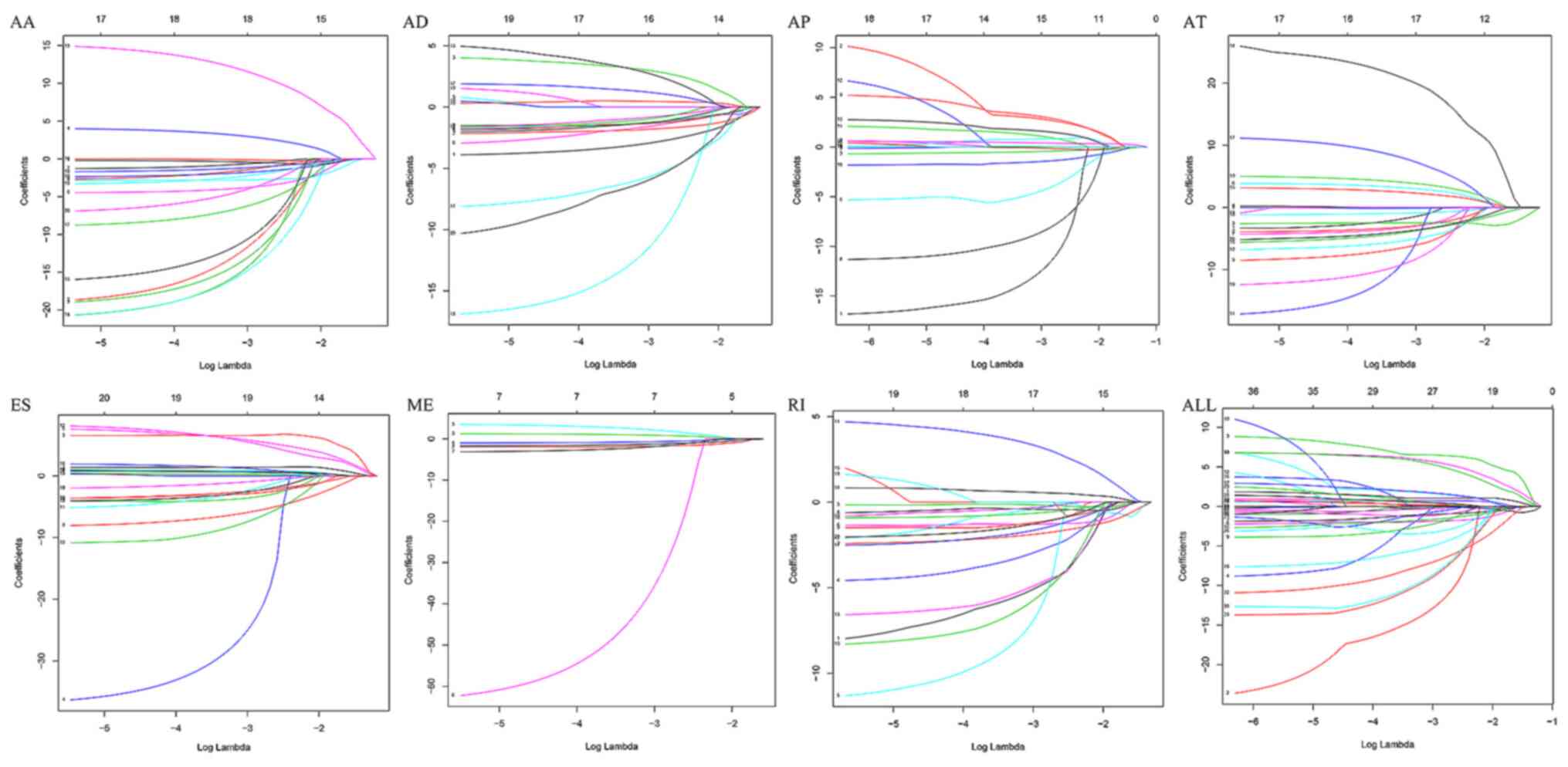
Least absolute shrinkage and selection operator
(LASSO) Cox analysis, which is ideal for high-dimensional data
(24), was performed to compute the
optimal coefficient and the deviance likelihood for each prognostic
feature using the ‘glmnet’ package in R (version 4.0–2; http://cran.r-project.org/web/packages/glmnet/index.html).
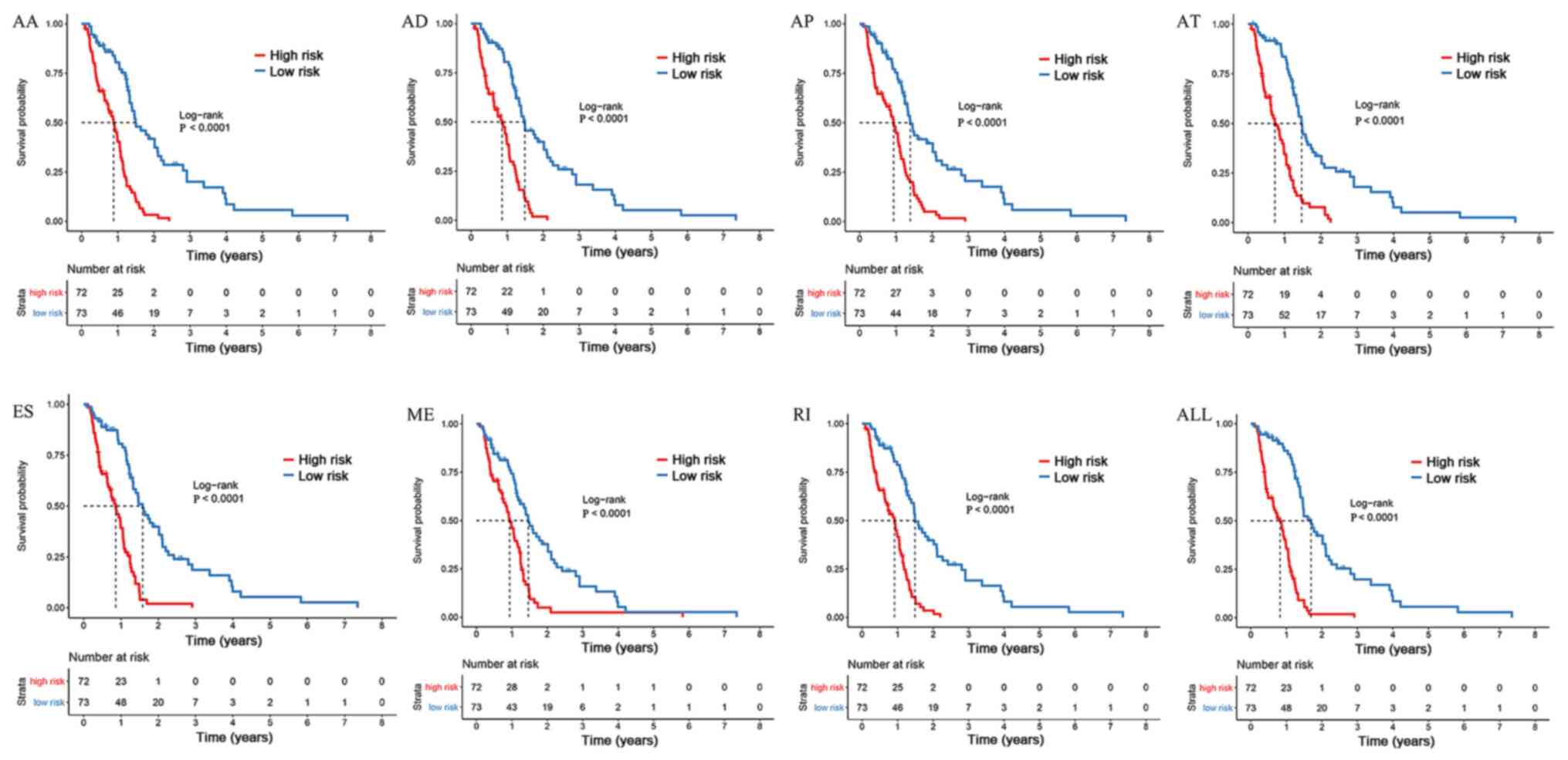
According to each coefficient, the AS events were divided into
high- and low-risk subgroups based on the median risk scores (All,
7.59; AA, 1.89; AD, 1.74; AP, 4.05; AT, 2.78; ES, 3.41; ME, 1.2;
RI, 3.03). Kaplan-Meier survival analysis and log-rank test were
used to further validate whether they resulted in diametrically
distinct outcomes. Prognostic models were calculated by multiplying
the PSI values of each significant splicing gene and the
coefficient performed by LASSO Cox analysis.
Clinical prognostic analyses
Clinical parameters were obtained to assess the
changes in AS events concerning the prognosis of patients with GBM.
A total of 169 GBM samples and 5 normal tissues with available
RNA-seq data were identified. Only cases with primary tumours, with
no adjuvant pre-operative therapy and with ≥30 days of OS were
included. There were 145 patients (51 females and 94 males; mean
age, 59.83; age range, 21–85 years) with applicable clinical
parameters and RNA-seq data who were included. To assess the
efficiency of each prognostic candidate, the survivalROC package in
R (version 1.0.3; http://cran.r-project.org/web/packages/survivalROC/index.html)
was used to generate the area under the curve (AUC) of receiver
operating characteristic (ROC) curves with censored data for each
model (25).
Evaluation of splicing-based prognostic signature as
an independent predictor was performed by integrating the following
clinical parameters into the univariate and multivariable Cox
regression analysis: Age, sex, IDH status, MGMT promoter status and
the risk score of AS events. The ability of the models to predict
the survival outcome of patients with GBM was evaluated. All
analyses were performed using R/Bioconductor (version 3.5.1;
http://www.r-project.org/).
Correlation between splicing events
and splicing factors
The correlation network between the gene expression
levels of SFs and the PSI values of AS events were performed by
Pearson's correlation analysis and plotted using Cytoscape (version
3.6.1; http://cytoscape.org/).
Results
Identification of survival-associated
AS events
mRNA-seq datasets and clinical information of
patients with GBM were obtained from TCGA (TCGA-GBM). A total of
169 GBM samples and 5 normal tissues with available RNA-seq data
were identified. Only cases with primary tumours, with no adjuvant
pre-operative therapy and with ≥30 days of OS were included. A
total of 145 patients with applicable clinical parameters and
RNA-seq data were finally included.
For evaluation of prognostic values (Table I and Fig.
1A), there were a total of 3,827 alternate acceptor (AA) events
in 2,684 genes, 3,269 AD events in 2,270 genes, 8,686 AP events in
3,476 genes, 8,456 AT events in 3,695 genes, 18,360 ES events in
6,935 genes, 184 ME events in 180 genes and 2,828 RI events in
1,897 genes. Hence, there may be ≥2 AS events in one gene
associated with survival in patients with GBM.
 | Table I.Summary of the glioblastoma sample
cohort from The Cancer Genome Atlas. |
Table I.
Summary of the glioblastoma sample
cohort from The Cancer Genome Atlas.
|
| Number of RNA-seq
events | Number of
survival-associated RNA-seq events |
|---|
|
|
|
|
|---|
| Splicing type | AS events | Genes | AS events | Genes |
|---|
| AA | 3,827 | 2,684 | 115 | 109 |
| AD | 3,269 | 2,270 | 110 | 106 |
| AP | 8,686 | 3,476 | 346 | 235 |
| AT | 8,456 | 3,695 | 264 | 179 |
| ES | 18,360 | 6,935 | 631 | 537 |
| ME | 184 | 180 | 7 | 7 |
| RI | 2,828 | 1,897 | 96 | 93 |
| ALL | 45,610 | 10,434 | 1,569 | 1,180 |
Based on the univariate Cox regression analysis, a
total of 115 AA events in 109 genes, 110 AD events in 106 genes,
346 AP events in 235 genes, 264 AT events in 179 genes, 631 ES
events in 537 genes, 7 ME events in 7 genes and 96 RI events in 93
genes were identified as significant prognosis-associated AS events
(P<0.05; Table I). Small nuclear
ribonucleoprotein-associated protein N, heterogeneous nuclear
ribonucleoprotein F, MRPS28, STAT3 and 26S proteasome non-ATPase
regulatory subunit 4 were considered as hub genes in the network
(Fig. 1B). Fig. 2A-G shows the top 20 significant
survival-associated AS events based on PSI values. The two-sided
red curves were obtained by the significant AS events in the
volcano plot (Fig. 2H and Table SI).
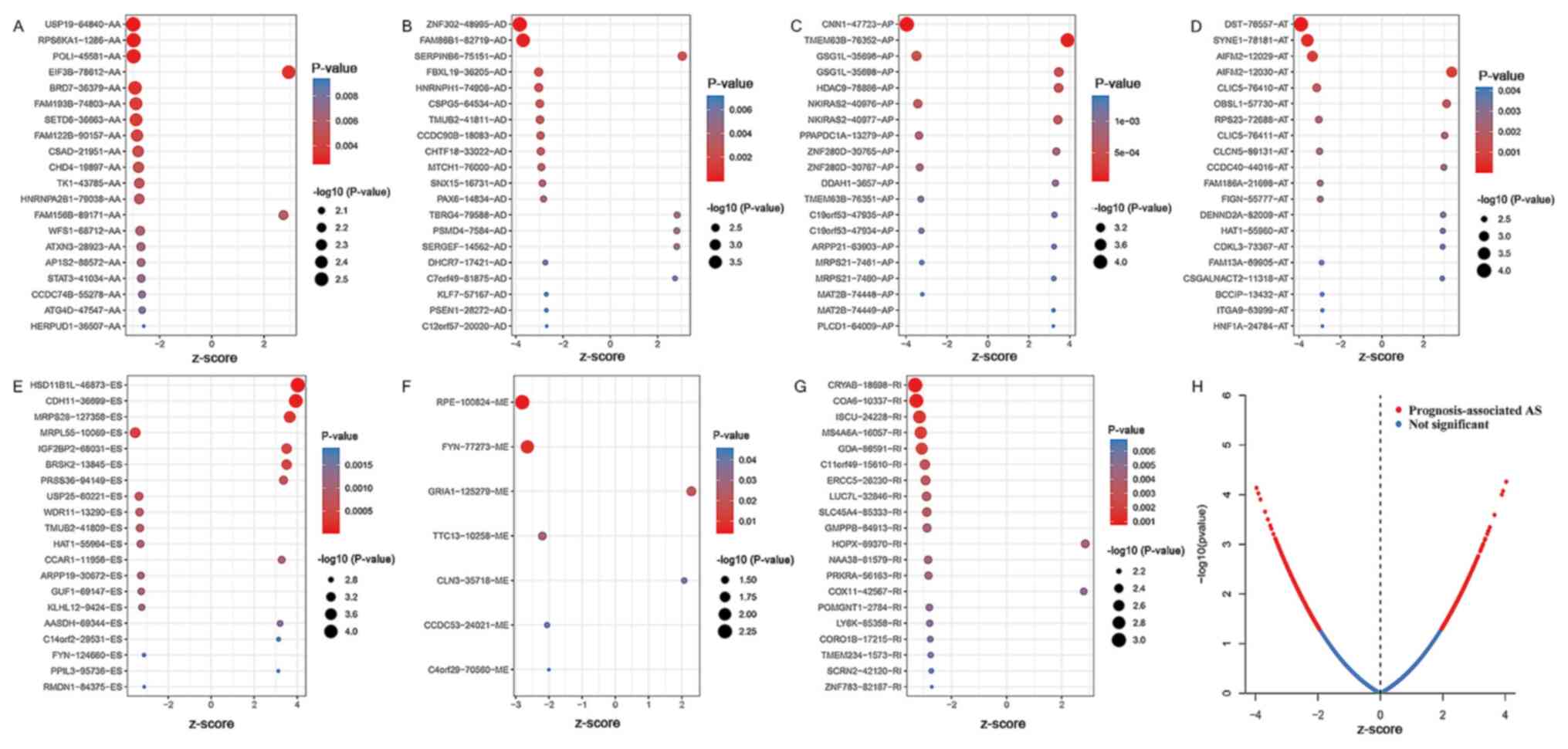 | Figure 2.Top AS events associated with overall
survival based on (A) AA, (B) AD, (C) AP, (D) AT, (E) ES, (F) ME
and (G) RI. The larger and more red dots indicate the alternative
splicing events with more significance. (H) Red dots represent
splicing events that are significantly prognosis-associated
(|z-score|>1). Blue dots represent splicing events without
prognosis association (|z-score|<1). The x-axis of the z-score
refers to either a positive or negative association. AS,
alternative splicing; AA, alternate acceptor; AD, alternate donor;
AP, alternate promoter; AT, alternate terminator; ES, exon
skipping; ME, mutually exclusive exon; RI, retained intron. |
Efficiency of prognostic models
Using LASSO Cox analysis, seven types of prognostic
models were developed based on AA, AD, AP, AT, ES, ME and RI
(Fig. 3). The risk score of each AS
type was calculated with their PSI values, while high- and low-risk
groups were divided using the median risk score as the cut-off
point. The present study revealed that the survival time of the
high-risk group was significantly shorter than that of the low-risk
group in the current cohort (Fig.
4). Therefore, the prognostic models of each AS type were
considered to predict the clinical outcome of patients with GBM
(Fig. 4). In addition, the 16 most
significant survival-associated AS events in the seven types were
selected to construct the final prognostic model (Table II). The scatter plots and heat maps
suggested that patients with high-risk scores had a low survival
time, while patients with low-risk scores had a high survival time
(Fig. S1). The final prognostic
model was deemed to be an ideal predictor of what could
significantly distinguish patients with GBM with distinct survival
times (Fig. S1). The AUC of the ROC
curve validated the performance of prognostic models with good
performance in prognosis prediction (Fig. 5). Additionally, univariate Cox
regression analysis was performed to assess the prognostic value of
clinical parameters, including age, sex, IDH mutation, MGMT
promoter methylation and risk score of AS events. Multivariate Cox
regression was applied after the sex parameter was eliminated since
there was no significant association between sex and survival in
the univariate analysis. The hazard ratios for risk score of AS
events were 1.0071 (95% CI, 1.0032–1.0111) and 1.0063 (95% CI,
1.0024–1.0103) by univariate and multivariate analyses,
respectively (Table III). In the
present cohort, the parameters of age, MGMT promoter methylation
status and risk score of AS events were considered as independent
factors of prognosis prediction.
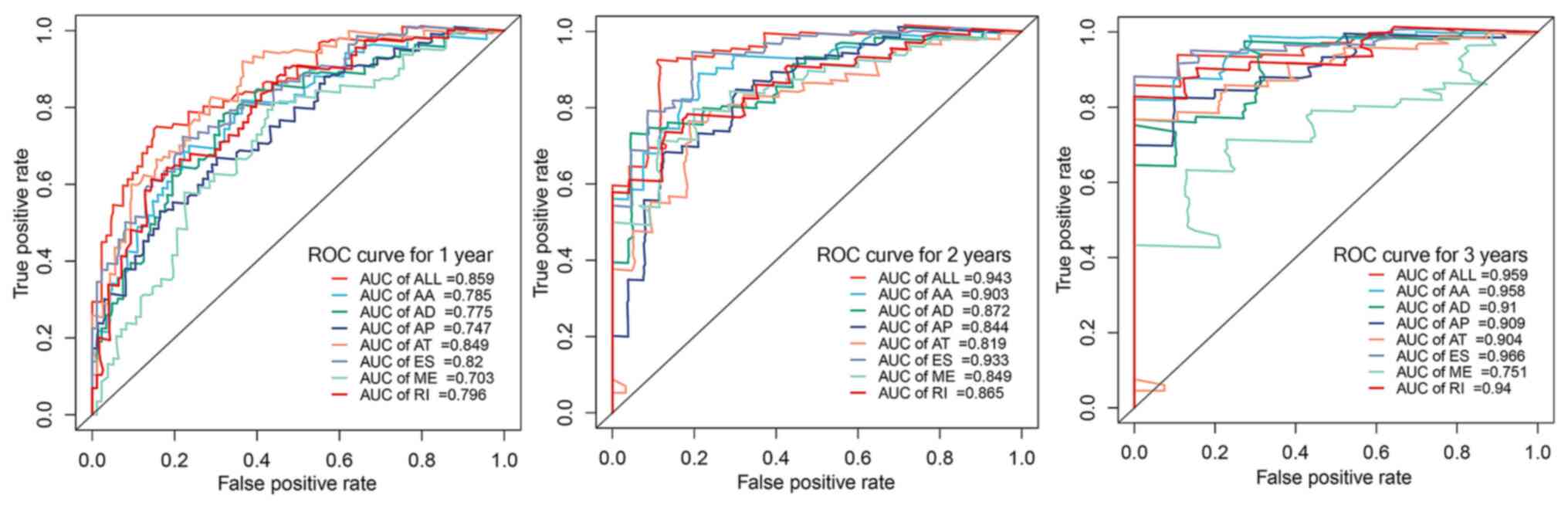 | Figure 5.AUC of ROC curves of eight types of
prognostic models for glioblastoma within 1, 2 and 3 years. AUC,
area under the curve; ROC, receiver operating characteristic; AA,
alternate acceptor; AD, alternate donor; AP, alternate promoter;
AT, alternate terminator; ES, exon skipping; ME, mutually exclusive
exon; RI, retained intron. |
 | Table II.Prognostic signatures based on each
type of alternative splicing event. |
Table II.
Prognostic signatures based on each
type of alternative splicing event.
| Splicing type | Algorithm | Hazard ratio (95%
CI) | AUC, 3 years |
|---|
| AA |
RPS6KA1|1286|AA*(−18.676)-POLI|45581|AA*21.795+EIF3B|78612|AA*
4.721-BRD7|36379|AA*4.239-FAM193B|74803|AA*5.174-FAM122B|90157|AA*2.925-CSAD|21951|AA*18.454-CHD4|19897|AA*2.244-TK1|43785|AA*24.736+FAM156B|89171|AA*16.546-ATXN3|28923|AA*14.630-STAT3|41034|AA*10.635-CCDC74B|55278|AA*1.472-ATG4D|47547|AA*3.458 | 3.755
(2.471–5.708) | 0.958 |
| AD |
ZNF302|48995|AD*(−3.8)+FAM86B1|82719|AD*2.205-FBXL19|36205|AD*
2.773-CSPG5|64534|AD*3.081-TMUB2|41811|AD*2.059-SNX15|16731|AD*9.014+TBRG4|79588|AD*5.12+C7orf49|81875|AD*1.91-KLF7|57167|AD*17.889-C12orf57|20020|AD*10.122-CCDC90B|18083|AD*2.252 | 3.824
(2.495–5.861) | 0.910 |
| AP |
CNN1|47723|AP*(−18.15)+TMEM63B|76352|AP*3.919-GSG1L|35696|AP*
1.009-NKIRAS2|40976|AP*9.203-PPAPDC1A|13279|AP*11.616+ZNF280D|30765|AP*5.021+
ARPP21|63903|AP*2.157 | 2.891
(1.933–4.323) | 0.909 |
| AT |
SYNE1|78181|AT*(−3.939)+OBSL1|57730|AT*5.011-RPS23|72688|AT*
4.493-CLCN5|89131|AT*7.954+CCDC40|44016|AT*5.905-FAM186A|21698|AT*18.776-FIGN|55777|AT*7.607+HAT1|55960|AT*30.067+CDKL3|73367|AT*3.414-FAM13A|69905|AT-6.427+CSGALNACT2|11318|AT*13.387-ITGA9|63999|AT*12.956-HNF1A|24784|AT*5.337 | 3.533
(2.353–5.306) | 0.904 |
| ES |
MRPL55|10069|ES*(−41.027)+BRSK2|13845|ES*8.338-USP25|60221|ES*
7.809-HAT1|55964|ES*10.427+CCAR1|11956|ES*11.785-ARPP19|30672|ES*6.302-GUF1|69147|ES*3.882-KLHL12|9424|ES*15.904+AASDH|69344|ES*2.443+C14orf2|29531|ES*1.304-FYN|124660|ES*
3.062+PPIL3|95736|ES*0.960 | 3.699
(2.433–5.623) | 0.966 |
| ME |
RPE|100824|ME*(−1.878)+GRIA1|125279|ME*1.332-TTC13|10258|ME*
1.255+CLN3|35718|ME*3.709-CCDC53|24021|ME*71.454-C4orf29|70560|ME*3.131 | 2.390
(1.612–3.544) | 0.751 |
| RI |
CRYAB|18698|RI*(−8.072)-COA6|10337|RI*2.972-MS4A6A|16057|RI*
5.522-GDA|86591|RI*10.733-SLC45A4|85333|RI*1.739-GMPPB|64913|RI*9.038+HOPX|69370|RI*4.469-PRKRA|56163|RI*7.561-LY6K|85358|RI*1.156-CORO1B|17215|RI*2.786-ZNF783|82187|RI*2.937 | 3.850
(2.519–5.885) | 0.940 |
| All |
HSD11B1L|46873|ES*1.533-CNN1|47723|AP*19.215+TMEM63B|76352|AP*2.915-ZNF302|48995|AD*2.545-FAM86B1|82719|AD*1.709-SYNE1|78181|AT*3.756+BRSK2|13845|ES*8.183-PPAPDC1A|13279|AP*6.446+PRSS36|94149|ES*1.304-USP25|60221|ES*11.622-CRYAB|18698|RI*5.015-HAT1|55964|ES*16.158+CCAR1|11956|ES*7.809+GUF1|69147|ES*2.621+ARPP21|63903|AP*2.198-KLHL12|9424|ES*-13.522 | 4.6097
(2.97–7.155) | 0.959 |
 | Table III.Cox regression analysis of clinical
parameters and risk score for assessing prognostic model value in
patients with glioblastoma. |
Table III.
Cox regression analysis of clinical
parameters and risk score for assessing prognostic model value in
patients with glioblastoma.
|
| Univariate Cox
regression | Multivariate Cox
regression |
|---|
|
|
|
|
|---|
| Clinical
variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, >60 vs. ≤60
years | 1.0397
(1.0210–1.0587) |
2.57×10−05 | 1.0344
(1.0125–1.0567) | 0.00189 |
| Sex, male vs.
female | 1.0198
(0.6541–1.5899) | 0.93117 | N/A | N/A |
| IDH mutation, yes
vs. no | 0.1836
(0.0573–0.5881) | 0.00432 | 0.4370
(0.1272–1.5019) | 0.18876 |
| MGMT promoter
methylation, yes vs. no | 0.4987
(0.3129–0.7950) | 0.00345 | 0.5253
(0.3276–0.8425) | 0.00755 |
| Risk score of AS
events, high vs. low | 1.0071
(1.0032–1.0111) | 0.00036 | 1.0063
(1.0024–1.0103) | 0.00153 |
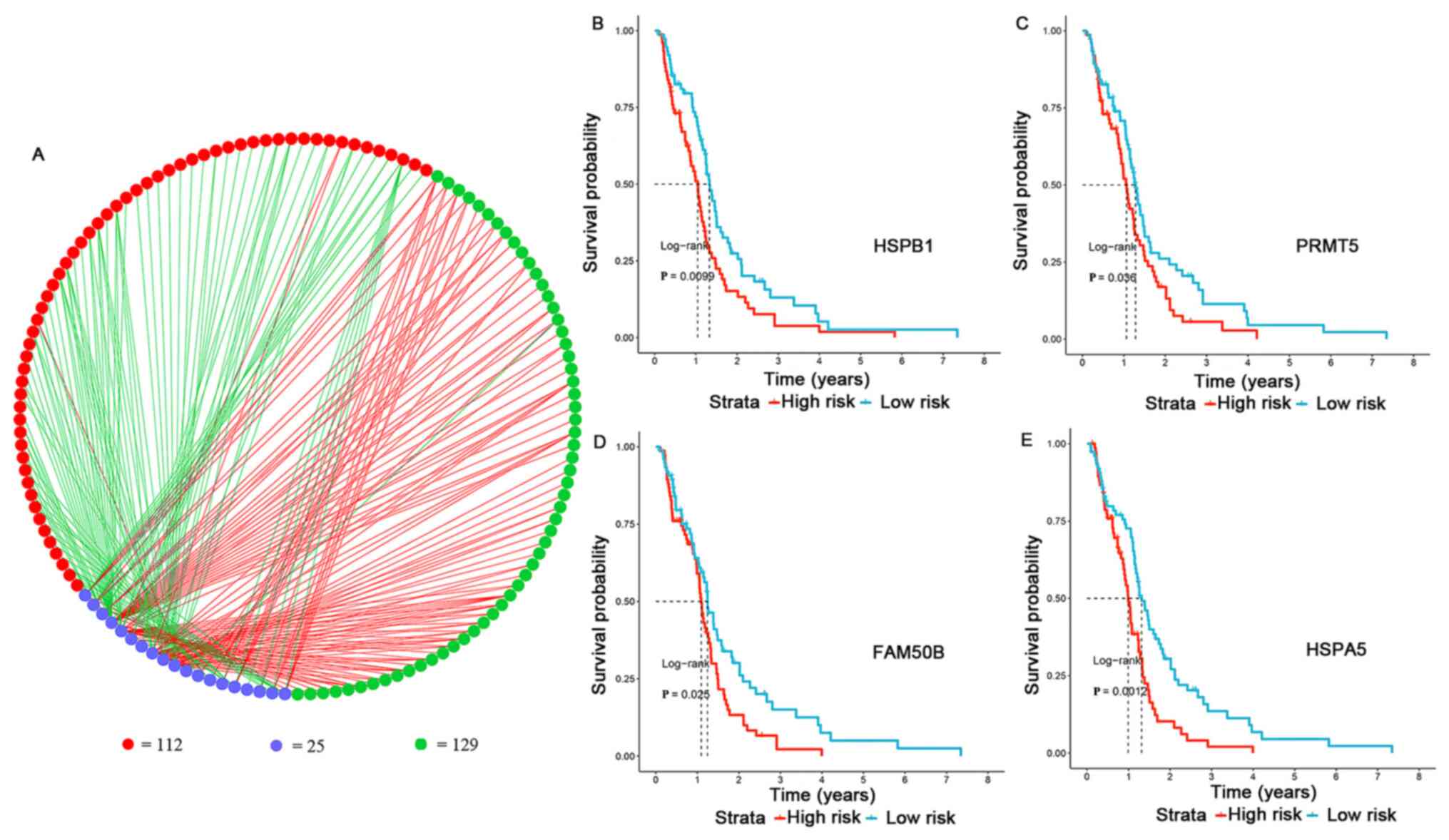
SF-AS regulatory association
The regulatory network of the significant
survival-associated AS events (n=241) and SFs was plotted using
Cytoscape (n=25) (Table SII and
Fig. 6A). Among all SFs, four
representative prognostic factors were selected, including heat
shock protein b-1 (HSPB1), protein arginine N-methyltransferase 5
(PRMT5), protein FAM50B (FAM50B) and endoplasmic reticulum
chaperone BiP (HSPA5). The expression levels of each SF gene were
calculated, while high- and low-risk groups were divided using the
median values as cut-off. The results revealed that the survival
time of the high-risk group was significantly shorter than that of
the low-risk group (P<0.05; Fig.
6B-E). Therefore, HSPB1, PRMT5, FAM50B and HSPA5 were
identified as significantly representative prognostic factors.
Among the splicing correlation network, a total of 129 favorable
prognosis AS events were correlated with survival-associated SFs,
while a total of 112 adverse prognosis AS events were correlated
with survival-associated SFs (P<1×10−10; Table SIII). Notably, the most favorable
splicing events were negatively regulated by SFs, while the most
adverse splicing events were positively regulated by SFs (Table SIII).
Discussion
GBM is a primary neuroepithelial tumour of the
central nervous system and accounts for 12–15% of all intracranial
tumours (1,2). The present study analyzed GBM datasets
composed of classical, proneural, mesenchymal and neural subtypes.
Patients with GBM have a poor median survival time of 12–15 months
following standard therapy, with only 3–5% of patients surviving up
to 5 years after the first diagnosis (1,2).
Currently, several molecular markers have been tested as part of
the routine clinical investigation of patients with GBM, including
MGMT, IDH, EGFR, PTEN, VEGF, TP53, p16INK4a gene and 1p19q gene, as
well as imaging biomarkers (26).
However, there is still a limited number of molecular signatures
for the contribution to anti-GBM therapies, such as temozolomide,
bevacizumab and lomustine (27).
Developments in next-generation sequencing methods have led to the
identification of specific molecular signatures of GBM that allow
for further investigation of the molecular pathogenesis of this
disease (28). In recent years,
high-throughput RNA-seq approaches have extensively promoted
genome-wide analyses, including genome splicing investigation. The
present study used bioinformatics techniques to identify
survival-associated AS events in order to construct splicing
signatures for the prediction of prognosis, orchestrate SF-AS
networks and assess their potential underlying molecular
mechanism.
Previously, SpliceSeq analyses have been adopted to
establish AS profiling and construct prognostic models in glioma;
several potential AS events were identified in pan-glioma and GBM
cohorts, including adenine DNA glycosylase, metalloreductase
STEAP3, SUMO-conjugating enzyme UBC9, von Hippel-Lindau disease
tumor suppressor, BTB/POZ domain-containing protein KCTD7, protein
S100-A4, endothelin-converting enzyme 2 and lymphocyte antigen 6K
(29–31). Additionally, several prognostic
models based on AS events have been constructed for prognosis
prediction, which may complement the molecular classification,
further identify potential glioma subgroups and highlight SFs as an
important mechanism of splicing regulation in the carcinogenesis
and aggressiveness of GBM (9,31–33).
The present study comprehensively analyzed the
prognostic value of AS events and SFs in a GBM cohort using several
computational approaches. The in-depth study further investigated
alterations of mRNA-seq for prognostic monitoring. The ideal
prognostic model built by combining all significant AS events
exhibited potential for predicting the survival outcome of patients
with GBM. Splicing correlation network analysis further revealed
regulated nodes, revealing the potential mechanisms in the
regulatory network at the genome-wide level.
In the interaction network analysis, HSPB1, PRMT5,
FAM50B and HSPA5 were identified as independent prognostic factors.
It has been reported that HSPB1 (also known as Hsp27)
phosphorylation leads to the activation of orphan nuclear receptor
TAK1 and TAK1-p38/ERK pro-survival signaling, thus acting against
TNF-α-induced apoptosis (34). PRMT5
is one of the candidate genes required for apoptosis or loss of
self-renewal for differentiated and undifferentiated GBM cells,
respectively (35). The specificity
and efficacy of four novel PRMT5 inhibitors have been identified
for the treatment of GBM (36).
Additionally, a previous study has validated that a family with
sequence similarity to FAM50B serves a key role as
methylation-based biomarkers for the diagnosis and treatment of GBM
(37). Furthermore, by specifically
inhibiting HSPA5, a new compound known as HA15 was able to increase
the unfolded protein response and lead to the death of cancer cells
by concomitant induction of autophagy and apoptosis, both in
vitro and in vivo (38).
Whether downregulation of specific SFs may affect the associated AS
events requires further validation in vivo.
Several limitations inevitably influenced the
reliability of the present results. There was a limited number of
patients with GBMs with complete clinicopathological parameters
recruited in the present analysis. All subtypes, including
classical, proneural, mesenchymal and neural, were analyzed without
precise classification. Therefore, subsequent functional
experiments in vitro and in vivo are required to
further validate the molecular mechanisms of how SFs regulate the
splicing process in GBM development.
Finally, the present study identified that
survival-associated AS events were favorable predictors and the
prognostic model performed well in predicting the stratification
for patients with GBM. According to these identified
survival-associated AS events and SFs, several valuable biomarkers
may be determined for further validation studies.
In conclusion, the present study established a
molecular phenomenon of OS-associated AS and SFs in patients with
GBM, which is valuable for investigating the underlying mechanisms
in the oncogenesis of GBM. The present findings may facilitate the
ongoing effort in developing novel transcriptome prognostic models
for the management of GBM. Further identification of prognostic SFs
and construction of an SF-AS network will advance the investigation
of splicing-associated mechanisms.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Shanghai
Municipal Commission of Health and Family Planning (grant no.
201640292).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The TCGA-GBM dataset generated and/or analyzed during the
current study is available in the TCGA repository (https://portal.gdc.cancer.gov/repository?facetTab=files&filters=%7B%22op%22%3A%22and%22%2C%22content%22%3A%5B%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22cases.primary_site%22%2C%22value%22%3A%5B%22brain%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22cases.project.program.name%22%2C%22value%22%3A%5B%22TCGA%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22cases.project.project_id%22%2C%22value%22%3A%5B%22TCGA-GBM%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22files.analysis.workflow_type%22%2C%22value%22%3A%5B%22HTSeq%20-%20FPKM%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22files.data_category%22%2C%22value%22%3A%5B%22transcriptome%20profiling%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22files.data_type%22%2C%22value%22%3A%5B%22Gene%20Expression%20Quantification%22%5D%7D%7D%5D%7D).
Authors' contributions
JQ, CW and YC contributed to the conception and
methodology of the study. CW, HH and XD contributed to the analysis
and acquisition of the data. JQ and SC wrote the manuscript and
contributed to the interpretation of the data. CW, SC and YC
supervised the study. YC acquired the funding. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15
(Suppl 2):ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Binabaj MM, Bahrami A, ShahidSales S,
Joodi M, Joudi Mashhad M, Hassanian SM, Anvari K and Avan A: The
prognostic value of MGMT promoter methylation in glioblastoma: A
meta-analysis of clinical trials. J Cell Physiol. 233:378–386.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao W, Li J, Chen MM, Luo Y, Ju Z, Nesser
NK, Johnson-Camacho K, Boniface CT, Lawrence Y, Pande NT, et al:
Large-scale characterization of drug responses of clinically
relevant proteins in cancer cell lines. Cancer Cell. Oct
2–2020.doi: 10.1016/j.ccell.2020.10.008 (Online ahead of print).
View Article : Google Scholar
|
|
5
|
Singh B and Eyras E: The role of
alternative splicing in cancer. Transcription. 8:91–98. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Y, Shi N, Regev A, He S and Hemann MT:
Integrated regulatory models for inference of subtype-specific
susceptibilities in glioblastoma. Mol Syst Biol. 16:e95062020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du J, Yan X, Mi S, Li Y, Ji H, Hou K, Ma
S, Ba Y, Zhou P, Chen L, et al: Identification of prognostic model
and biomarkers for cancer stem cell characteristics in glioblastoma
by network analysis of multi-omics data and stemness indices. Front
Cell Dev Biol. 8:5589612020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ,
Yan W, Liu YW, Zhang W, Chen L and Jiang T: Prognostic value of a
nine-gene signature in glioma patients based on mRNA expression
profiling. CNS Neurosci Ther. 20:112–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yin W, Tang G, Zhou Q, Cao Y, Li H, Fu X,
Wu Z and Jiang X: Expression profile analysis identifies a novel
five-gene signature to improve prognosis prediction of
glioblastoma. Front Genet. 10:4192019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carpenter S, Ricci EP, Mercier BC, Moore
MJ and Fitzgerald KA: Post-transcriptional regulation of gene
expression in innate immunity. Nat Rev Immunol. 14:361–376. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Antonopoulou E and Ladomery M: Targeting
splicing in prostate cancer. Int J Mol Sci. 19:12872018. View Article : Google Scholar
|
|
13
|
Kim HK, Pham MHC, Ko KS, Rhee BD and Han
J: Alternative splicing isoforms in health and disease. Pflugers
Arch. 470:995–1016. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brosseau JP, Lucier JF, Nwilati H,
Thibault P, Garneau D, Gendron D, Durand M, Couture S, Lapointe E,
Prinos P, et al: Tumor microenvironment-associated modifications of
alternative splicing. RNA. 20:189–201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dvinge H, Kim E, Abdel-Wahab O and Bradley
RK: RNA splicing factors as oncoproteins and tumour suppressors.
Nat Rev Cancer. 16:413–430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee Y and Rio DC: Mechanisms and
regulation of alternative Pre-mRNA splicing. Annu Rev Biochem.
84:291–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martinez-Montiel N, Rosas-Murrieta NH,
Anaya Ruiz M, Monjaraz-Guzman E and Martinez-Contreras R:
Alternative splicing as a target for cancer treatment. Int J Mol
Sci. 19:5452018. View Article : Google Scholar
|
|
18
|
Suñé-Pou M, Prieto-Sánchez S,
Boyero-Corral S, Moreno-Castro C, El Yousfi Y, Suñé-Negre JM,
Hernández-Munain C and Suñé C: Targeting splicing in the treatment
of human disease. Genes (Basel). 8:872017. View Article : Google Scholar
|
|
19
|
Noushmehr H, Weisenberger DJ, Diefes K,
Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP,
Bhat KP, et al: Identification of a CpG island methylator phenotype
that defines a distinct subgroup of glioma. Cancer Cell.
17:510–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res.
44:D1018–D1022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ryan MC, Cleland J, Kim R, Wong WC and
Weinstein JN: SpliceSeq: A resource for analysis and visualization
of RNA-Seq data on alternative splicing and its functional impacts.
Bioinformatics. 28:2385–2387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lex A, Gehlenborg N, Strobelt H, Vuillemot
R and Pfister H: UpSet: Visualization of intersecting sets. IEEE
Trans Vis Comput Graph. 20:1983–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu G, Feng X and Stein L: A human
functional protein interaction network and its application to
cancer data analysis. Genome Biol. 11:R532010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szopa W, Burley TA, Kramer-Marek G and
Kaspera W: Diagnostic and therapeutic biomarkers in glioblastoma:
Current status and future perspectives. Bio Res Int.
2017:80135752017.
|
|
27
|
Taal W, Oosterkamp HM, Walenkamp AM,
Dubbink HJ, Beerepoot LV, Hanse MC, Buter J, Honkoop AH, Boerman D,
de Vos FY, et al: Single-agent bevacizumab or lomustine versus a
combination of bevacizumab plus lomustine in patients with
recurrent glioblastoma (BELOB trial): A randomised controlled phase
2 trial. Lancet Oncol. 15:943–953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aldape K, Zadeh G, Mansouri S,
Reifenberger G and von Deimling A: Glioblastoma: Pathology,
molecular mechanisms and markers. Acta Neuropathol. 129:829–848.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeng Y, Zhang P, Wang X, Wang K, Zhou M,
Long H, Lin J, Wu Z, Gao L and Song Y: Identification of prognostic
signatures. 2020.
|
|
30
|
Xie ZC, Wu HY, Dang YW and Chen G: Role of
alternative splicing signatures in the prognosis of glioblastoma.
Cancer Med. 8:7623–7636. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Zhao C, Guo B, Zhao Z, Wang H and
Fang Z: Systematic profiling of alternative mRNA splicing signature
for predicting glioblastoma prognosis. Front Oncol. 9:9282019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Y, Ren Z, Peng Y, Li K, Wang X, Huang
G, Qi S and Liu Y: Classification of glioma based on prognostic
alternative splicing. BMC Med Genomics. 12:1652019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J and Manley JL: Misregulation of
pre-mRNA alternative splicing in cancer. Cancer Discov.
3:1228–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qi Z, Shen L, Zhou H, Jiang Y, Lan L, Luo
L and Yin Z: Phosphorylation of heat shock protein 27 antagonizes
TNF-α induced HeLa cell apoptosis via regulating TAK1
ubiquitination and activation of p38 and ERK signaling. Cell
Signal. 26:1616–1625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Banasavadi-Siddegowda YK, Russell L, Frair
E, Karkhanis VA, Relation T, Yoo JY, Zhang J, Sif S, Imitola J,
Baiocchi R and Kaur B: PRMT5-PTEN molecular pathway regulates
senescence and self-renewal of primary glioblastoma neurosphere
cells. Oncogene. 36:263–274. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Banasavadi-Siddegowda YK, Welker AM, An M,
Yang X, Zhou W, Shi G, Imitola J, Li C, Hsu S, Wang J, et al: PRMT5
as a druggable target for glioblastoma therapy. Neuro Oncol.
20:753–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jia D, Lin W, Tang H, Cheng Y, Xu K, He Y,
Geng W and Dai Q: Integrative analysis of DNA methylation and gene
expression to identify key epigenetic genes in glioblastoma. Aging
(Albany NY). 11:5579–5592. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cerezo M and Rocchi S: New anti-cancer
molecules targeting HSPA5/BIP to induce endoplasmic reticulum
stress, autophagy and apoptosis. Autophagy. 13:216–217. 2017.
View Article : Google Scholar : PubMed/NCBI
|