Introduction
The fine-tuning of the intracellular levels of
reactive oxygen species (ROS) and the cellular redox state is of
crucial importance for breast carcinogenesis, progression,
prognosis and therapy (1). ROS are
involved in multiple redox regulated signaling pathways including
the modulation of endoplasmic reticulum and mitochondria
homeostasis and contribute to the pathogenesis of a number of human
diseases including breast cancer (2). Cancer cells generate persistently high
ROS levels which activate pro-survival mechanisms including
endoplasmic reticulum (ER) stress, unfolded protein response (UPR)
and inhibition of pro-apoptotic pathways initiated in mitochondria
(1,3). ER stress is activated when the ER
chaperones involved in the refolding of misfolded proteins are not
functional leading to the accumulation of high levels of misfolded
proteins in the cell and subsequent induction of UPR (4). By activating ER stress, the cells
transiently halt protein synthesis in order to cope with the
accumulation of non-functional proteins but if the concentration of
the misfolded proteins is high the UPR stimulates cell death
(5,6).
Redox responsive endoplasmic reticulum proteins such
as members of the protein disulfide isomerase (PDI) superfamily and
in particular the prototype family member prolyl 4-hydroxylase
subunit beta (P4HB or PDIA1) activates the ER transmembrane kinase
protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK)
thus playing a crucial role in the stimulation of the UPR (7). PDIA1 has been shown to modulate
cellular oxidative stress mediating homeostasis of the antioxidant
glutathione (8) and its function is
regulated by cellular redox state. Specifically, PDIA1 exerts
chaperone and isomerase activities in an oxidative stress-dependent
manner facilitating redox-mediated regulation of protein folding
(9–12).
PDIA1 is primarily localized in the endoplasmic
reticulum but extracellular matrix, mitochondria, nucleus, and
cytosolic PDIA1 localization has also been demonstrated (13). Diverse subcellular localization
confers PDIA1 a dual role in cell death and survival during ER
stress (14). For instance, PDIA1
mitochondrial localization contributes to the induction of
mitochondrial outer membrane permeabilization (MOMP), the release
of the mitochondrial cytochrome c into the cytoplasm and the
initiation of apoptosis (15).
Furthermore, PDIA1 mitochondrial localization suggested that this
protein is involved in the regulation of redox mediated
mitochondrial functions including energy metabolism (16). Nuclear localization of PDIA1
implicated this protein in the regulation of the activity of
redox-sensitive transcription factors to co-ordinate efficient ROS
detoxification (17). Extracellular
matrix localization of PDIA1 was potentially related to invasion
and metastasis as it has been shown for PDI family members in
glioma (18) and hepatocarcinoma
(14).
The impact of ER stress on immune responses
(19) as well as the involvement of
the UPR in the regulation of the tumor microenvironment are
becoming widely acknowledged (20).
PDI family members are key components of the antigen presentation
machinery (21,22) contributing to the selection and
loading of the optimal antigen to MHC class I molecules (23,24)
and the dissociation of the antigen-loaded MHC class I complex and
exit from the endoplasmic reticulum (25).
In the present study, the regulation of the PDIA1
functions under diverse oxidative stress conditions and its role in
a range of physiological processes known to affect breast
carcinogenesis were investigated in the estrogen receptor
(ERα)-positive MCF-7 and the triple negative MDA-MB-231 breast
cancer cells. The correlation between PDIA1 and HLA-G
mRNA levels and their association with the overall survival were
also investigated in breast cancer patients by analyzing the
molecular taxonomy of breast cancer international consortium
(METABRIC) microarray dataset available in the cBio cancer genomics
portal (http://cbioportal.org) (26).
The results provide evidence to support the view
that PDIA1 differentially affects antioxidant homeostasis
and ATP generation in the ERα-positive vs. the TNBC cells and its
mRNA levels are linked to the overall survival of stage 2 breast
cancer patients. The findings of this study indicate an alternative
molecular mechanism directing the evasion of immune surveillance in
breast tumors that could be used as a platform for the design of
stratified breast cancer immunotherapies, in addition to those that
use PDIA1 as a therapeutic target (27).
Materials and methods
Cell culture
The human breast carcinoma cell lines MCF-7
(expressing wild-type p53) and MDA-MB-231 [bearing mutated p53
(R280K)] were obtained from the European collection of cell
cultures (ECACC) and maintained in Dulbeccos modified Eagles medium
(Sigma-Aldrich, UK) supplemented with 10% foetal bovine serum
(Gibco) and 1% penicillin/streptomycin (Lonza) at 37°C in a
humidified atmosphere containing 5% CO2 until they
reached 70% confluency (48 h). Where indicated cells were treated
with 10 ng/ml interferon-γ (IFN-γ) (Sigma-Aldrich) for 24 h or 10
µM etoposide (ETOP) for 24 h (Sigma-Aldrich).
Western blotting
Cellular extracts from MCF-7 and MDA-MB-231 were
collected in ice-cold TNN buffer containing 1 mM DTT, 1 mM PMSF,
and 1 µg/ml protease inhibitors. Protein concentrations were
determined by the Bradford assay (Sigma Aldrich) and 30 µg of
protein per sample were resolved on a 20% precast polyacrylamide
gel and transferred to a PVDF membrane. The membranes were blocked
using 5% fat-free milk in PBS (v/v) for 1 h at 25°C. Membranes were
then incubated in 2.5% milk in PBS-0.1% Tween-20 (v/v) with
anti-P4HB antibody (Santa Cruz Biotechnology; sc-136230) (dilution
1:500) or β-actin (Sigma Aldrich; A1978) (dilution 1:10,000)
overnight at 4°C. The membranes were washed three times with
PBS-0.1% Tween-20 (v/v) for 5 min and then incubated with secondary
anti-mouse immunoglobulin G conjugated to horsedish peroxidase (GE
Healthcare) (dilution 1:1,000) in 2.5% milk in PBS-0.1% Tween-20
(v/v) for 1 h at 25°C. Protein bands were then visualized using the
ChemiDoc MP imaging system (Bio-Rad).
siRNA transfection
A concentration of 5 µM of the siGENOME P4HB siRNA
and 5 µM of the siGENOME non-targeting siRNA pool was added to each
well containing 2×105 cells in DMEM and incubated for 72
h according to the suppliers instructions (Dharmacon) as described
previously (28). Immediately after
the 72 h transfection cells were used for subsequent
experimentation. The sequences of the siRNA pools against P4HB and
scramble siRNA were: P4HB: ACAGGACGGUCAUUGAUUA,
GGACGGUCAUUGAUUACAA, CCAAGAGUGUGUCUGACUA, CAGAGAGGAUCACAGAGUU and
scramble: UAGCGACUAAACACAUCAA, UAAGGCUAUGAAGAGAUAC,
AUGUAUUGGCCUGUAUUAG, AUGAACGUGAAUUGCUCAA.
Intracellular ROS generation
ROS generation was measured using the dye
CM-H2DCF-DA (29).
Briefly, cells were grown in 6-well plates at a density of
1×106 cells/well and treated as indicated. Cells were
centrifuged for 3 min at 400 × g at 25°C, the supernatant was
removed and the pellet resuspended in cold PBS. Then, 1 µM of
CM-H2DCF-DA was added and the extracts were incubated
for 30 min in the dark at 25°C. Subsequently, 30 µl of samples were
transferred to A2 slides (ChemoMetec) and ROS were measured using
the NucleoCounter NC-3000™ (ChemoMetec).
Glutathione cellular levels
The NucleoCounter NC-3000™ system was used to detect
changes in the intracellular level of (reduced) thiols. Following
the indicated treatments cells were dissociated from the culture
plates using 500 µl dissociation buffer (ChemoMetec). Then the
cells were stained with solution 5 according to the manufacturers
instructions (ChemoMetec), loaded onto 8-chamber NC-slides and
samples were analysed using the NucleoCounter NC-3,000™.
Mitochondrial membrane disruption
Mitochondrial transmembrane disruption was measured
using the cationic dye JC-1 (5,5,6,6-tetrachloro-1,1,3,3-tetraethyl
benzimidazol carbocyanine iodide) (ChemoMetec) and the
NucleoCounter NC-3,000™. Following the indicated treatment, the
cells were stained with JC-1 and DAPI according to the
manufacturers instructions (ChemoMetec). Cellular JC-1 monomers and
aggregates were detected as green and red fluorescence,
respectively. Mitochondrial depolarization and apoptosis were
revealed as a decrease in the ratio of red/green fluorescence.
Staining with the blue fluorescent dye (DAPI) was used to detect
necrotic and late apoptotic cells. After stained cells were loaded
on an 8-chamber NC-Slide A8™ and samples were analysed quantifying
the amount of blue, green, and red fluorescence using the
NucleoCounter NC-3,000™.
Adenosine triphosphate assay
ATP levels were measured using the ViaLight plus kit
(Lonza), based on the amount of bioluminescent ATP generated in
cells. ATP monitoring reagent (AMR plus) was prepared by adding
assay buffer into the vial containing the lyophilized AMR and
incubated at room temperature for 15 min for complete rehydration.
Cells were lysed in 50 µl and the cell lysate was added to a
luminometer plate together with 100 µl of cell lysis reagent for 10
min. A total volume of 100 µl of AMR plus was added to the
appropriate well. The plate was then incubated at room temperature
for 2 min and luminescence values were obtained using the Fluostar
OPTIMA microplate reader (BMG Labtech).
HLA-G surface levels
Breast cancer cell lines were seeded in a 6-well
plate at a concentration of 1×106 cancer cells/well for
24 h in a CO2 incubator. Cells were then transfected
with P4HB siRNA or scramble siRNA for 48 h at 37°C. Following
treatment with IFN-γ or ETOP for 24 h at 37°C, the cells were
detached from the wells with 500 µl dissociation buffer and the
cell lysates were transferred to a 15 ml falcon tube and
centrifuged at 300 × g for 5 min at 4°C. The supernatant was
removed and 3×105 cells in 100 µl (10% FBS in PBS) were
transferred to a fresh tube. APC-anti human HLA-G (Thermo Fisher)
was added and the cells were incubated for 30 min on ice in the
dark. The stained samples were analysed by flow cytometry (Becton
Dickinson).
PDIA1-HLA-G correlation in breast
cancer patients
The METABRIC breast cancer dataset was downloaded
from cBio Cancer Genomics Portal (http://cbioportal.org) for 1,900 breast cancer
patients (26). The distribution of
the mRNA expression levels of PDIA1 and HLA-G was
measured across samples. PDIA1 (P4HB) and
HLA-G mRNA expression levels were considered high if their
z-scores were higher or equal to the 75th percentile of the
distribution (30) and low if their
z-scores were lower than the 75th percentile of the distribution
(z-score high PDIA ≥ 0.572, z-score high HLA-G≥
0.517).
Statistical analysis
Graphs were plotted using GraphPad PRISM 7 (GraphPad
software Inc.). The Mann-Whitney test was used to assess the
significance of the difference between two groups, whereas the
Kruskal-Wallis test was used to assess the significance of the
differences between more than two groups. Log-rank was used to test
the significance of the difference between Kaplan-Meier survival
curves. The mean was calculated as ± SEM. P<0.05 indicated
statistical significance.
Results
Silencing of PDIA1 gene expression in
breast cancer cells
To investigate the role of the PDIA1 in the
modulation of diverse hallmarks of cancer such as resistance to
cell death, regulation of cellular energy production pathways and
evasion of immune destruction, the gene expression of this
oxidoreductase was silenced in the ERα-positive (MCF-7) and the
ERα-negative (MDA-MB-231) breast cancer cells. The efficiency of
silencing of the PDIA1 gene expression was confirmed for
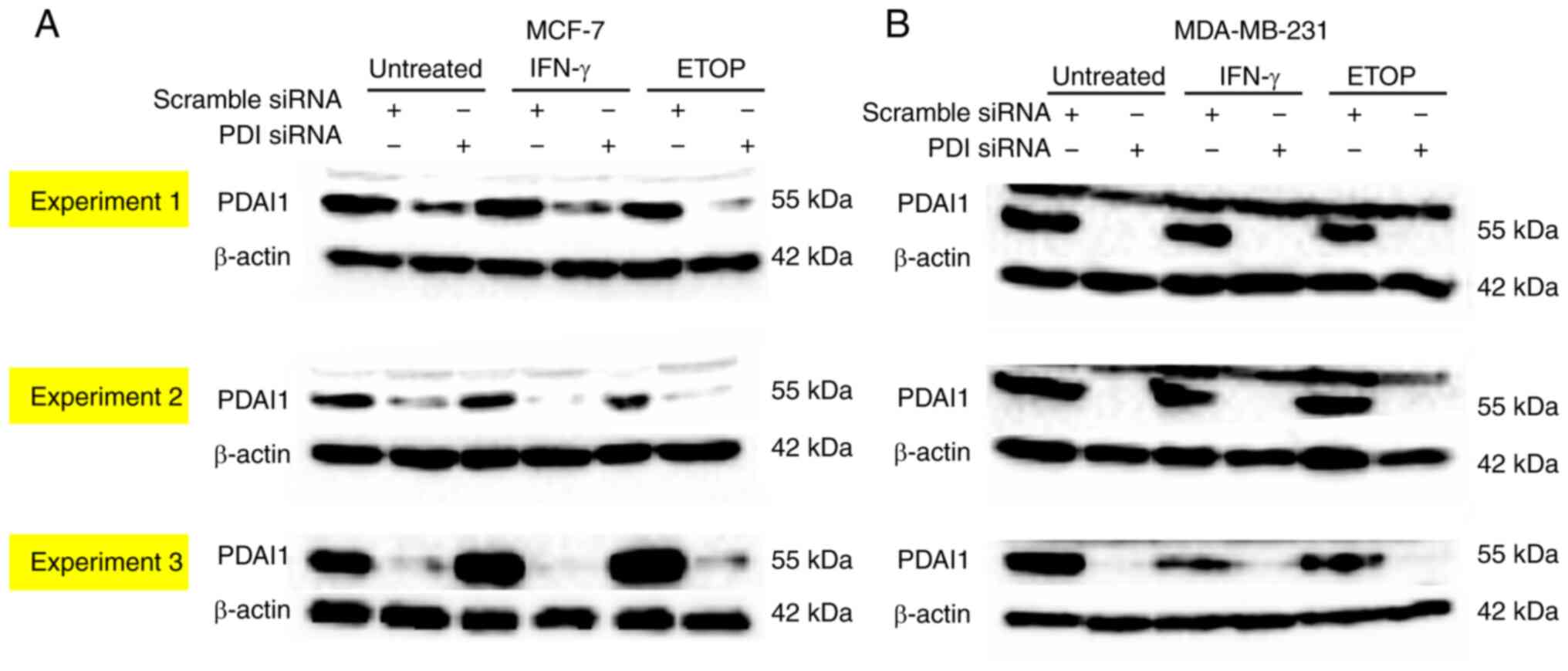
each assay performed. The representative western blot analysis
shown in Fig. 1, which shows the
efficient silencing of the PDIA1 gene expression only in
MCF-7 and MDA-MD-231 cells transfected with the siRNA-PDIA1
untreated or treated with IFN-γ or ETOP, but not in those cells
transfected with the scramble siRNA.
PDIA1 modulates ROS generation in
breast cancer cells
Type II interferon IFN-γ induces both carcinogenic
and cytotoxic effects (31) playing
an important role in cell growth and survival in a manner involving
the generation of ROS (32). The
topoisomerase II inhibitor ETOP has also been shown to induce ROS
generation in brain cancer cells (33). Additionally, IFN-γ regulates the
expression of genes encoding NADPH oxidases (Nox) (34) and evidence indicating functional
interaction between PDI and Nox family members (35) suggests that IFN-γ may indirectly
regulate PDIA1 levels and activity under diverse oxidative stress
conditions. The association between overexpression of NADPH
oxidases and resistance to ETOP treatment as a result of
ROS-mediated induction of senescence has also been reported
(36), suggesting that PDIA1 is a
potential indirect modulator of ROS mediated effects exerted upon
ETOP treatment. To investigate the potential involvement of PDIA1
in altering the IFN-γ and ETOP mediated redox state in breast
cancer cells ROS levels were measured in MCF-7 and MDA-MB-231 cells
treated with these compounds in the presence or absence of
PDIA1.
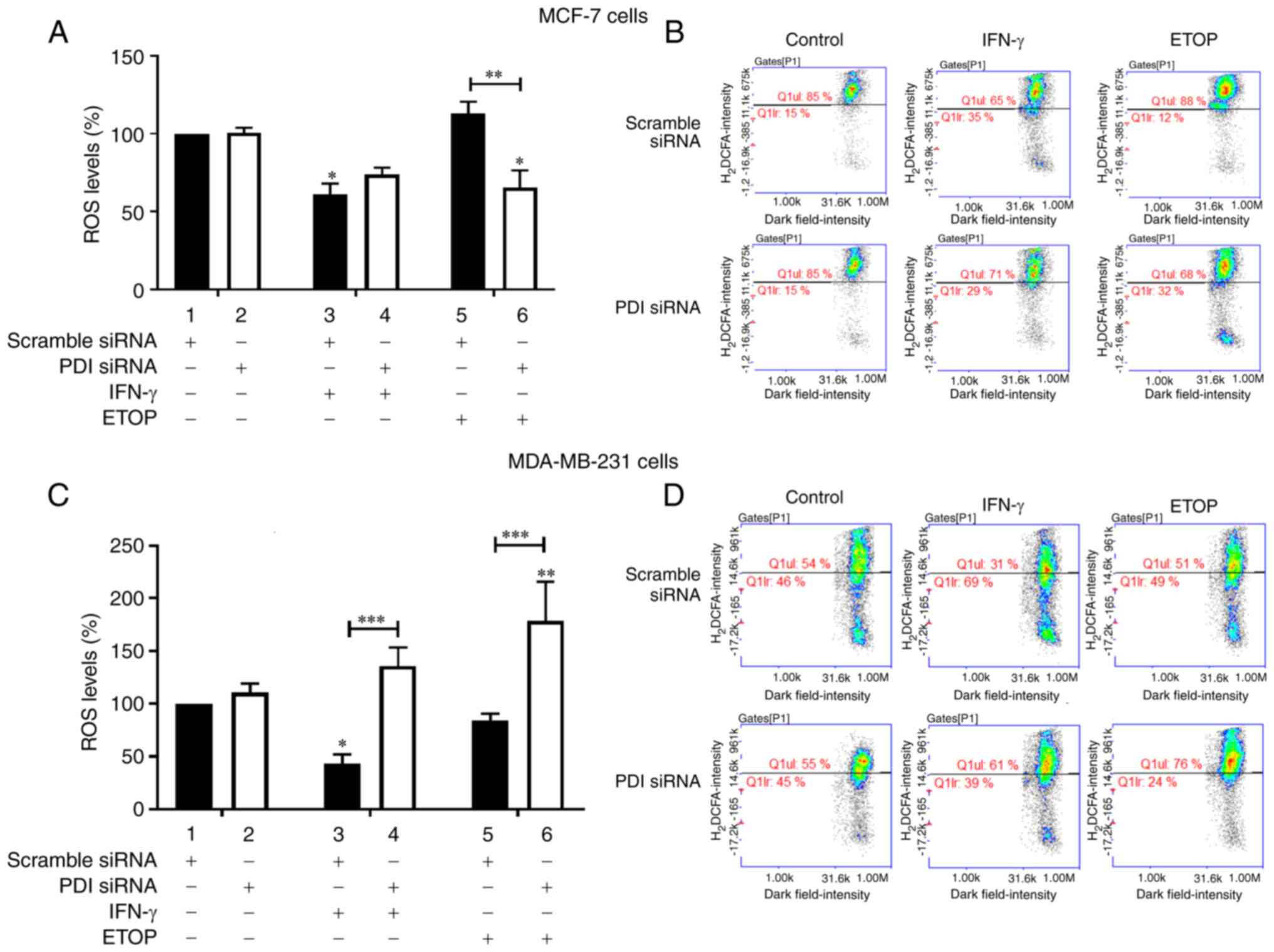
In MCF-7 cells silencing of PDIA1 did not affect ROS
generation in the untreated or IFN-γ treated cells compared to
scramble-transfected cells (Fig.
2A, compare bars 1 and 3 to bars 2 and 4, respectively).
Decreased ROS levels were recorded in PDIA1-silenced MCF-7 cells
treated with ETOP compared to scramble-transfected cells under the
same conditions (Fig. 2A, compare
bar 5 to bar 6). Decreased ROS levels were measured in MCF-7 cells
treated with IFN-γ in the presence of PDIA1 compared to the
untreated cells (Fig. 2A, compare
bar 3 to bar 1) whereas in the presence of PDIA1 ETOP did not
affect ROS generation in MCF-7 cells (Fig. 2A compare bar 5 to bar 1). In the
absence of PDIA1 significantly decreased ROS generation was
observed in the ETOP-treated MCF-7 cells compared to untreated
cells (Fig. 2A, compare bar 6 to
bar 2). Significantly increased ROS levels were observed in
PDIA1-silenced MDA-MB-231 cells treated with either IFN-γ or ETOP
compared to the scramble-transfected cells under the same
conditions (Fig. 2C, compare bars 3
and 5 to bars 4 and 6, respectively). Significantly decreased ROS
levels were observed in the IFN-γ treated MDA-MB-231 cells in the
presence of PDIA1 compared to untreated cells under the same
conditions (Fig. 2C, compare bar 3
to bar 1). ETOP treatment of PDIA1-silenced MDA-MB-231 cells
resulted in a significant increase of ROS generation compared to
untreated cells (Fig. 2C, compare
bar 6 to bar 2). PDIA1 protein levels were not significantly
affected by any treatment (Fig. 1).
Representative histograms showing the ROS levels in scramble or
siRNA-PDIA1-transfected MCF-7 or MDA-MB-231 cells untreated or
treated with IFN-γ or ETOP are provided in Fig. 2B and 2D, respectively.
PDIA1 regulates GSH levels in breast
cancer cells
The ratio of glutathione (GSH) vs. glutathione
disulfide (GSSG) is important for cellular physiology since a
decreased GSH/GSSG ratio leads to increased susceptibility to
oxidative stress whereas increased GSH levels induce resistance of
cancer cells to oxidative stress (37). Inhibition of PDIA1 isomerase
activity has been shown to abrogate glutathione depletion (8) indicating the essential role of PDIA1
in the process of disulfide bond formation and therefore the
regulation of the ratio of GSH vs. GSSG (38). To investigate the role of PDIA1 in
the regulation of the GSH homeostasis in breast cancer cells the
GSH/GSSG ratio was investigated in MCF-7 and MDA-MB-231 cells in
which the PDIA1 expression had been silenced.
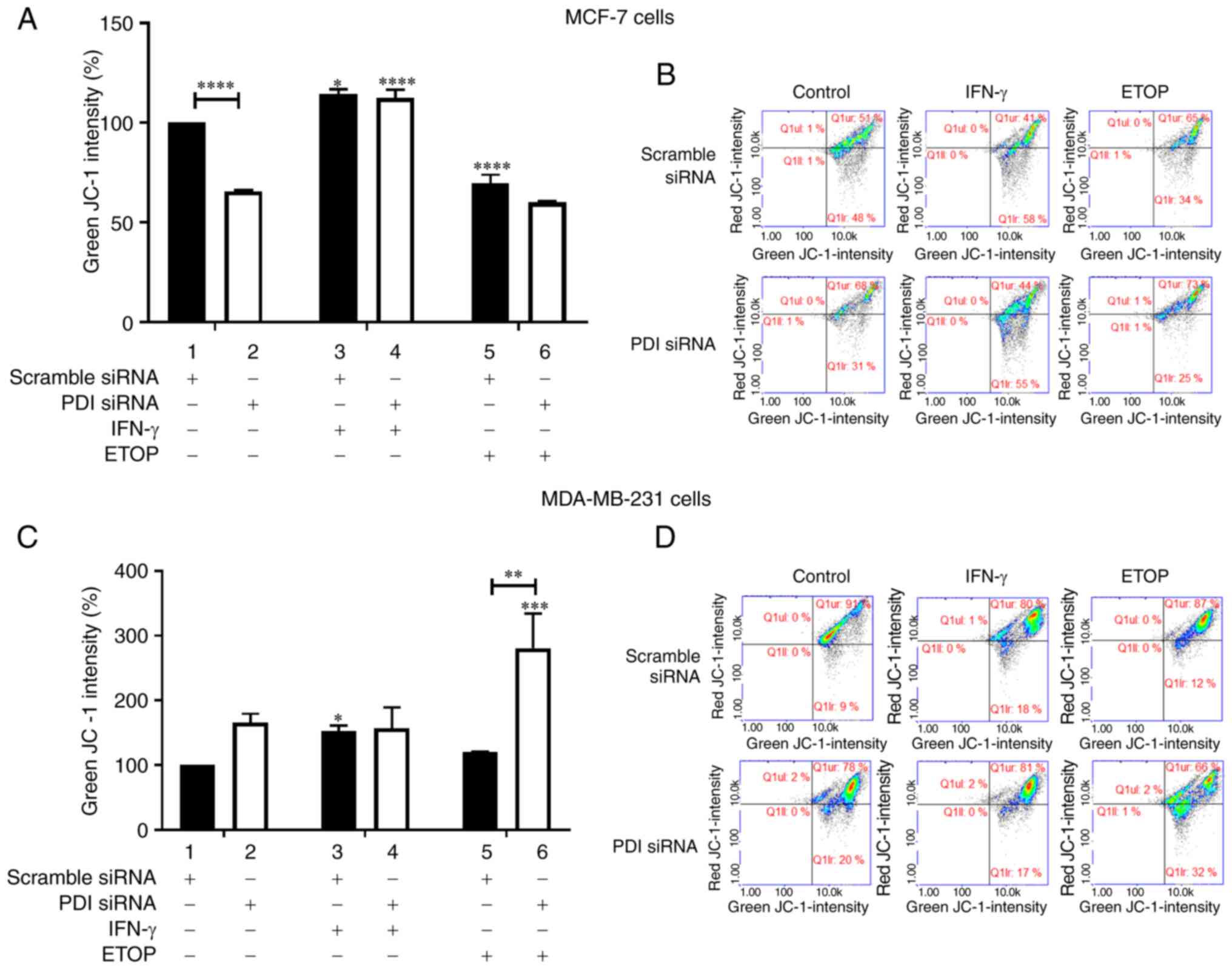
Significant upregulation of GSH levels was observed
in the untreated PDIA1-silenced MCF-7 cells compared to
scramble-transfected cells under the same conditions (Fig. 3A, compare bar 1 to bar 2). There
were no other changes evident in GSH levels in MCF-7 cells in any
of the conditions studied (Fig.
3A). By contrast, in the MDA-MB-231 cells significant
downregulation of GSH levels was observed in the untreated
PDIA1-silenced MDA-MB-231 cells compared to the untreated
and scramble-transfected cells (Fig.
3C, compare bar 1 to bar 2). Downregulation of GSH levels was
also recorded in the ETOP-treated MDA-MB-231 cells in the presence
of PDIA1 compared to the untreated cells (Fig. 3C, compare 5 to bar 1). Finally
upregulated GSH levels were measured in PDIA1 silenced
MDA-MB-231 cells treated with IFN-γ compared to the siRNA-PDIA1
transfected untreated cells (Fig.
3C, compare bar 4 to bar 2). Representative histograms showing
the GSH levels in scramble or siRNA-PDIA1 transfected MCF-7 or
MDA-MB-231 are provided in Fig. 3B
and 3D respectively.
PDIA1 regulates mitochondrial function
in breast cancer cells
The communication between ER and mitochondria is
critical for several important cellular functions including the
balance between survival and death (39). The role of PDIA1 in the regulation
of this communication has been shown in cells in which blocking the
PDIA1 activity prevents stimulation of the mitochondrial outer
membrane potential (MOMP) and inhibits apoptosis (15). In addition, the combination of ETOP
with a PDI inhibitor has been shown to trigger increased
ER-associated calcium influx and loss of mitochondrial membrane
integrity (40). These findings led
us to investigate the effect of PDIA1 on the integrity of the
mitochondrial membrane of breast cancer cells.
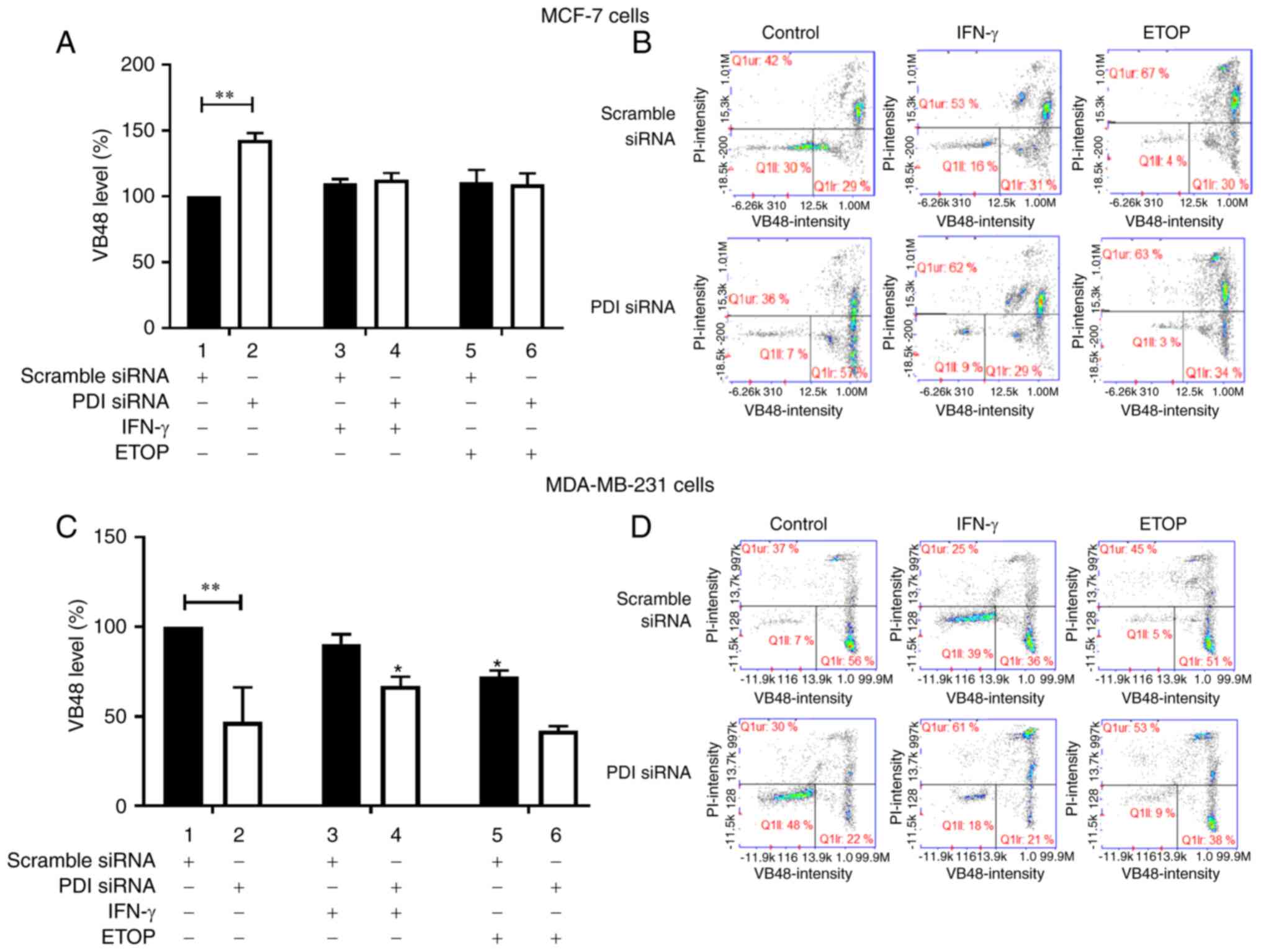
The mitochondrial membrane potential was
investigated in MCF-7 and MDA-MB-231 cells in which the
PDIA1 expression had been silenced and cells were left
untreated or treated with IFN-γ or ETOP. Silencing of PDIA1
in MCF-7 cells downregulated mitochondrial membrane potential in
the untreated cells compared to the scramble-transfected cells
under the same conditions (Fig. 4A,
compare bar 1 to bar 2). Increased mitochondrial membrane potential
was observed in the presence of PDIA1 in MCF-7 cells treated with
IFN-γ compared to the untreated cells (Fig. 4A, compare bar 3 to bar 1), whereas
decreased mitochondrial membrane disruption was measured in the
presence of PDIA1 in MCF-7 cells treated with ETOP compared
to the untreated cells (Fig. 4A,
compare bar 5 to bar 1). PDIA1-silenced MCF-7 cells treated
with IFN-γ exhibited increased mitochondrial membrane disruption
compared to PDIA1-silenced untreated cells (Fig. 4A, compare bar 4 to bar 2). Silencing
of PDIA1 in MDA-MB-231 cells upregulated mitochondrial
membrane disruption in ETOP-treated cells compared to the
scramble-transfected cells under the same conditions (Fig. 4C, compare bar 5 to bar 6). Increased
mitochondrial membrane disruption levels were also measured in
MDA-MB-231 cells treated with IFN-γ in the presence of PDIA1
compared to the untreated cells (Fig.
4C, compare bar 3 to bar 1). In ETOP-treated
PDIA1-silenced MDA-MB-231 cells the mitochondrial membrane
disruption was upregulated compared to the untreated cells under
these conditions (Fig. 4C, compare
bar 6 to bar 2). Representative histograms showing the intensity of
JC-1 (%) in scramble or siRNA targeting PDIA1-transfected MCF-7 and
MDA-MB-231 cells are provided in Fig.
4B and 4D, respectively.
PDIA1 modulates energy metabolism in
breast cancer cells
The communication between ER and mitochondria by a
network of proteins residing in the interface between the two
organelles is critical in controlling vital physiological functions
including energy metabolism and cellular death/survival decisions
(41). Structural and functional
changes of the ER-mitochondria contact proteins result in the
deregulation of calcium (Ca2+) homeostasis and
consequently mitochondrial energy production and cell death
(42). PDI plays an important role
in the cross talk between cellular redox state and Ca2+
homeostasis in the ER (43)
suggesting its involvement in the regulation of energy metabolism
(44).
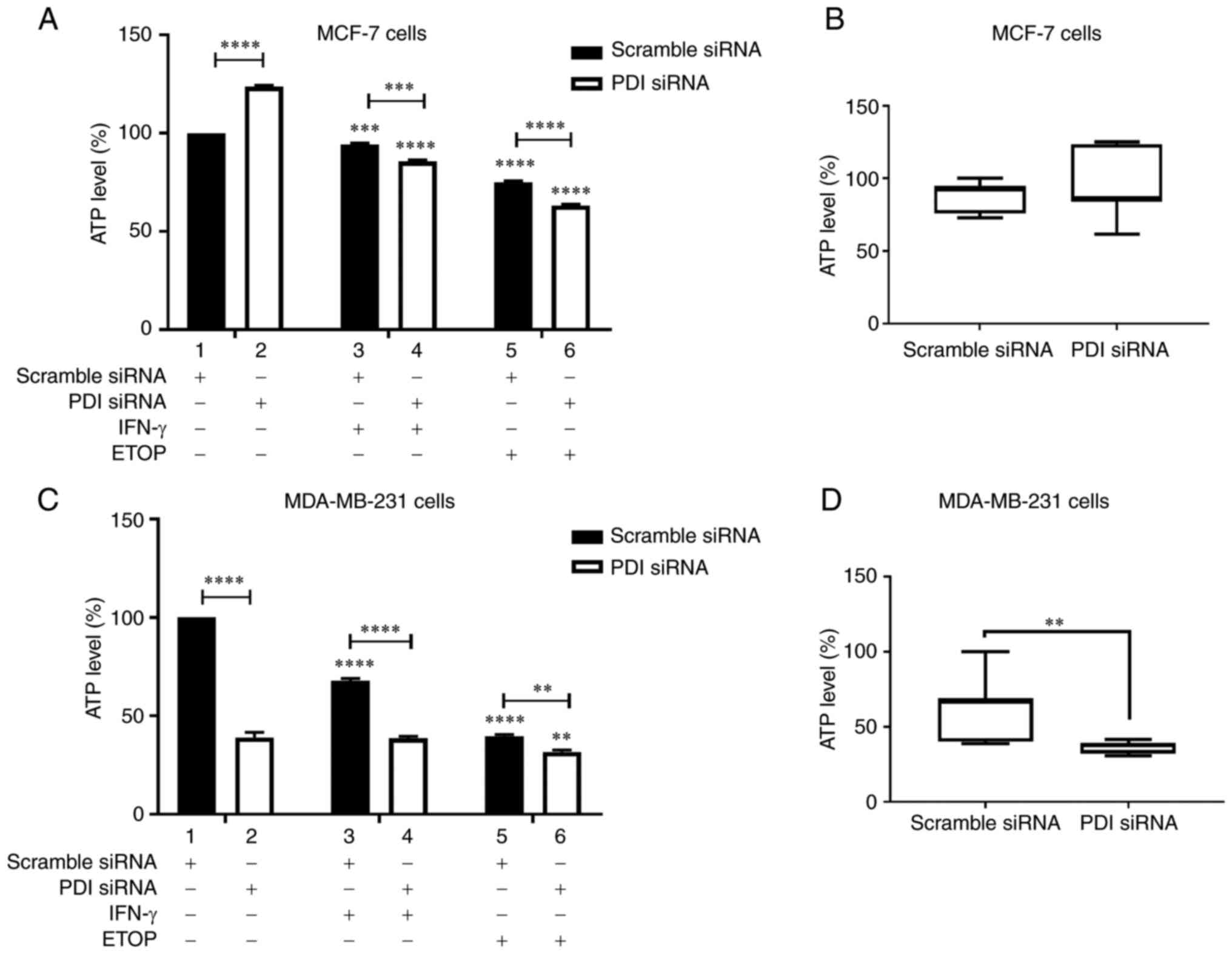
To test this hypothesis ATP production was
investigated in MCF-7 and MDA-MB-231 cells treated with IFN-γ or
ETOP in the presence or absence of PDIA1 (Fig. 5). Silencing of PDIA1 in MCF-7 cells
upregulated ATP levels in the untreated cells (Fig. 5A, compare bar 1 to bar 2). Decreased
ATP levels were recorded in MCF-7 cells treated with IFN-γ or ETOP
in the presence of PDIA1 compared to the untreated cells (Fig. 5A, compare bars 3 and 5 to bar 1).
Decreased ATP levels were also observed in PDIA1-silenced MCF-7
cells treated with IFN-γ (Fig. 5A,
compare bar 3 to bar 4) or ETOP compared to scramble-transfected
cells under the same conditions (Fig.
5A, compare bar 5 to bar 6). Overall, no significant changes in
ATP levels were evident in untreated, IFN-γ- or ETOP-treated
PDIA1-silenced MCF-7 cells compared to those measured in the
untreated, IFN-γ or ETOP treated cells in the presence of
PDIA1 (Fig. 5B)
Decreased ATP levels were measured in MDA-MB-231
cells treated with IFN-γ or ETOP in the presence of PDIA1
compared to the untreated cells (Fig.
5C, compare bars 3 and 5 to bar 1). Silencing of PDIA1
in MDA-MB-231 cells downregulated ATP levels in the untreated cells
(Fig. 5C, compare bar 1 to bar 2),
IFN-γ-treated cells (Fig. 5C,
compare bar 3 to bar 4), and ETOP-treated cells (Fig. 5C, compare bar 5 to bar 6). ATP
generation was downregulated significantly in untreated, IFN-γ- or
ETOP-treated MDA-MB-231 cells in the absence of PDIA1 vs.
the presence of PDIA1 (Fig.
5D).
PDIA1 modulates HLA-G surface levels
in breast cancer cells
In recent years, the connection between ER stress
signalling pathways, UPR induction, deregulation of energy
metabolism, and immune responses has been unravelled (45,46).
The link between the cellular redox state with several ER-resident
chaperones and in particular PDIA1 in mediating the processing,
optimal selection and antigen loading to the MHC class I during the
process of antigen presentation have been demonstrated (23,24,47).
Apart from the peptide antigen processing, loading and
stabilization of the early MHC class I complex (22), PDIA1 has also been shown to modulate
the MHC class I expression (48).
Two types of the MHC class I molecules have been described; the
classical (human leukocyte antigen HLA-A, HLA-B, and HLA-C alleles)
and the non-classical (HLA-E, HLA-F, HLA-G) proteins (49). Human breast cancer tissues and
breast cancer cells express HLA-G whereas in normal epithelial
mammary cells HLA-G mRNA expression has not been detected
indicating the involvement of the non-classical MHC class I
molecules in the evasion of the immune surveillance by breast
cancer cells and their important role in determining the prognosis
of breast cancer patients (50–52).
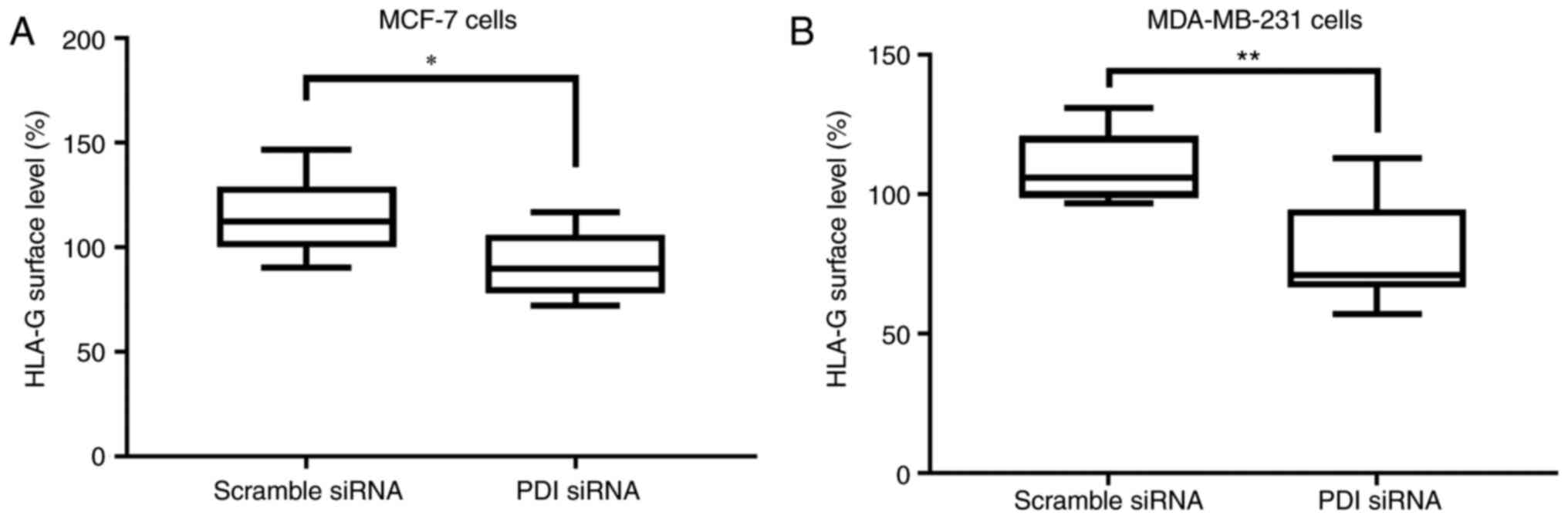
To investigate any potential role of PDIA1 in
modulating the surface expression of the non-classical MHC class I
HLA-G molecule and thus in the process of evasion of immune
surveillance (51,52) the HLA-G surface levels were followed
in untreated MCF-7 and MDA-MB-231 cells or treated with IFN-γ or
ETOP in the presence or absence of PDIA1 (Fig. 6). Significantly lower HLA-G surface
levels were observed in both MCF-7 and MDA-MB-231 cells in the
absence of PDIA1 compared to MCF-7 and MDA-MB-231 cells in
the presence of PDIA1 (Fig. 6A
and B).
PDIA1/HLA-G mRNA ratio correlates with
overall survival in breast cancer patients
To explore the correlation between PDIA1 and
HLA-G gene expression and identify any potential clinical
implications of the PDIA1 and HLA-G ratio in breast
cancer patient data obtained from the METABRIC dataset available in
the cBio Cancer Genomics Portal (http://cbioportal.org) (26) were analyzed to follow the
PDIA1 and HLA-G mRNA levels in various stages of
ERα-positive and ERα-negative breast cancer patients. Table I indicates the number of
ERα-positive and ERα-negative patients and the stage classification
of their disease. Results shown in Fig. S1 indicate the PDIA1 and
HLA-G mRNA expression levels in ERα-positive (Fig. S1A and B) and ERα-negative (Fig. S1C and D) patients in each stage of
the disease. The correlation between PDIA1 and HLA-G
mRNA levels in ERα-positive stage 1 (Fig. S2A), stage 2 (Fig. S2B) and stage 3 (Fig. S2C) patients indicates statistically
significant results for stage 2 and 3 patients. A similar analysis
of the correlation between PDIA1 and HLA-G mRNA
levels in ERα-negative stage 1 (Fig.
S2D), stage 2 (Fig. S2E) and
stage 3 (Fig. S2F) patients did
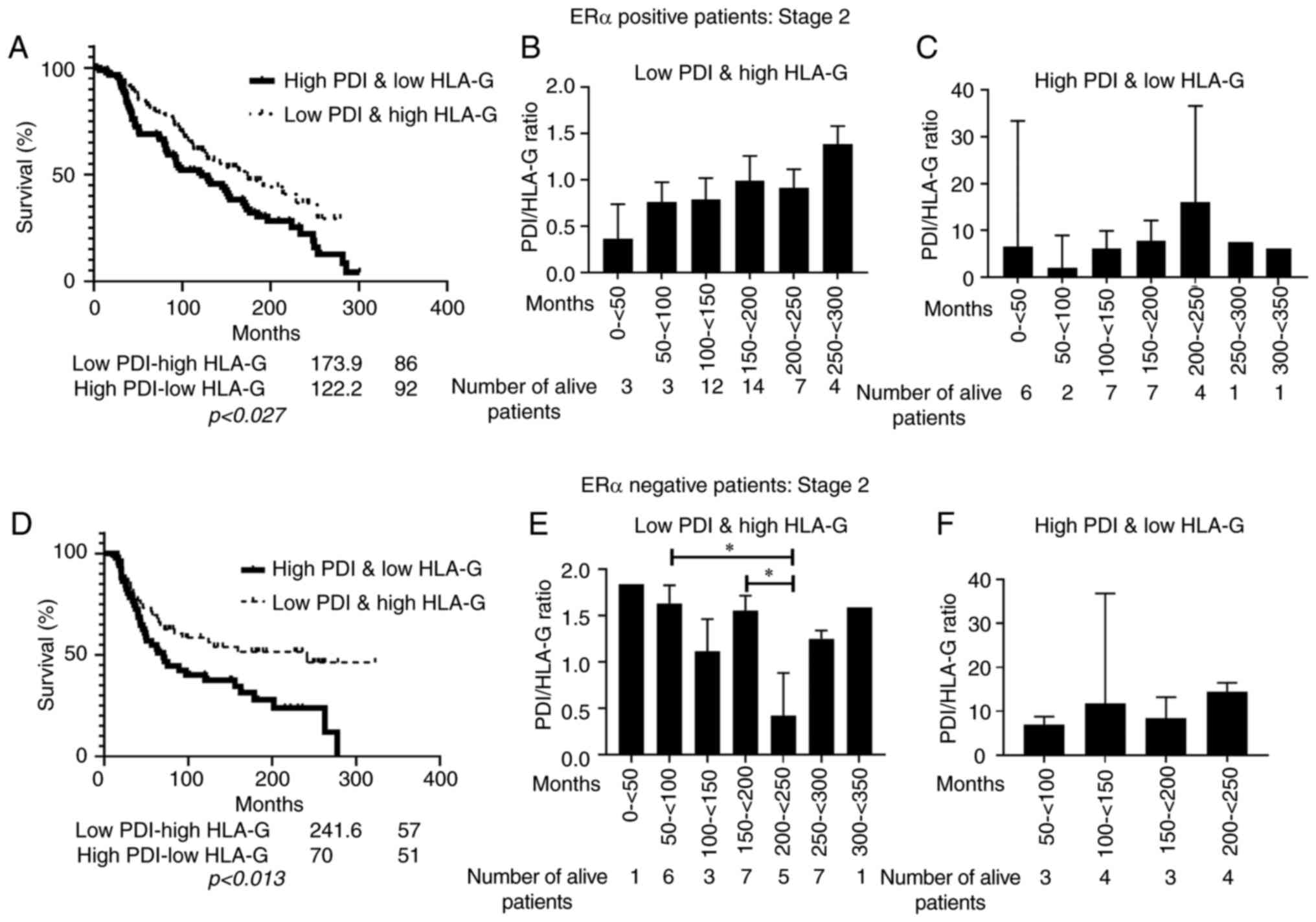
not show any statistical significance. Kaplan-Meir survival curves
were plotted to explore whether the PDIA1/HLA-G mRNA ratio
was associated with the overall survival of stage 2 breast cancer
patients (Fig. 7). Statistically
significant association between low PDIA1/high HLA-G mRNA ratio and
longer overall survival was observed in ERα-negative stage 2
patients (Fig. 7D). Analysis of the
subgroup of the living breast cancer patients at stage 2 exhibited
low PDIA1 and high HLA-G mRNA ratio indicated that in the
ERα-positive patients longer survival is associated predominantly
with high PDIA1 and low HLA-G mRNA levels (Fig. 7B) whereas in the ERα-negative
patients longer survival is associated mainly with low PDIA1
and high HLA-G mRNA levels (Fig.
7E). A similar analysis carried out in the subgroup of the
living breast cancer patients at stage 2 exhibiting high PDIA1 and
low HLA-G mRNA levels indicated that longer survival was associated
mainly with high PDIA1 and low HLA-G mRNA levels in
the ERα-negative patients (Fig. 7F)
but there was no correlation in the ERα-positive patients (Fig. 7C).
 | Table I.Breast cancer patient data obtained
from the METABRIC microarray dataset categorized by ERα status,
stage of the disease and levels of PDIA1 and HLA-G mRNA expression
levels. |
Table I.
Breast cancer patient data obtained
from the METABRIC microarray dataset categorized by ERα status,
stage of the disease and levels of PDIA1 and HLA-G mRNA expression
levels.
| METABRIC |
|---|
| ERα-positive |
| Not known |
| 378 | 1,457 |
|
| Stage 1 | Low PDIA1 | Low HLA-G | 265 |
|
|
|
|
| High HLA-G | 59 |
|
|
|
| HighPDIA1 | Low HLA-G | 48 |
|
|
|
|
| High HLA-G | 15 |
|
|
| Stage 2 | Low PDIA1 | Low HLA-G | 399 |
|
|
|
|
| High HLA-G | 86 |
|
|
|
| HighPDIA1 | Low HLA-G | 92 |
|
|
|
|
| High HLA-G | 38 |
|
|
| Stage 3 | Low PDIA1 | Low HLA-G | 38 |
|
|
|
|
| High HLA-G | 15 |
|
|
|
| HighPDIA1 | Low HLA-G | 14 |
|
|
|
|
| High HLA-G | 1 |
|
|
| Stage 4 | Low PDIA1 | Low HLA-G | 6 |
|
|
|
|
| High HLA-G | 1 |
|
|
|
| HighPDIA1 | Low HLA-G | 2 |
|
|
|
|
| High HLA-G | 0 |
|
| ERα-negative |
| Not known |
| 123 | 443 |
|
| Stage 1 | Low PDIA1 | Low HLA-G | 27 |
|
|
|
|
| High HLA-G | 24 |
|
|
|
| HighPDIA1 | Low HLA-G | 21 |
|
|
|
|
| High HLA-G | 16 |
|
|
| Stage 2 | Low PDIA1 | Low HLA-G | 45 |
|
|
|
|
| High HLA-G | 57 |
|
|
|
| HighPDIA1 | Low HLA-G | 51 |
|
|
|
|
| High HLA-G | 32 |
|
|
| Stage 3 | Low PDIA1 | Low HLA-G | 18 |
|
|
|
|
| High HLA-G | 13 |
|
|
|
| HighPDIA1 | Low HLA-G | 10 |
|
|
|
|
| High HLA-G | 6 |
|
|
| Stage 4 | Low PDIA1 | Low HLA-G | 0 |
|
|
|
|
| High HLA-G | 0 |
|
|
|
| HighPDIA1 | Low HLA-G | 0 |
|
|
|
|
| High HLA-G | 0 |
|
| Total |
|
|
|
| 1,900 |
Discussion
Protein disulfide isomerases compose a superfamily
of more than 20 members of endoplasmic reticulum proteins that
apart from protein folding exert multiple other functions including
oxidoreductase activity, molecular chaperoning and acting as
hormone reservoirs (10). The
prototype member of the family PDIA1 is localized primarily in the
endoplasmic reticulum but nuclear, mitochondrial and localization
on the surface of the cellular membrane has also been reported
(43). PDIA1 exerts
tumor-stimulating or suppressing effects being involved in a wide
spectrum of physiological functions in a manner dependent on the
type of tissue, microenvironmental conditions, subcellular
localization and its oxidized or reduced conformation (10–12).
To shed light on the role of the PDIA1 in breast carcinogenesis we
followed a variety of ROS modulated pathways in the estrogen
receptor positive MCF-7 and the ERα negative MDA-MB-231 cells under
differential oxidative stress conditions in the presence or absence
of PDIA1.
In the present study, IFN-γ treatment reduced ROS
levels in both MCF-7 and MDA-MB-231 cells whereas ETOP did not
affect ROS generation in these cells in the presence of PDIA1. In
the absence of PDIA1 IFN-γ treatment did not affect ROS levels in
MCF-7 cells and increased oxidative stress in MDA-MB-231 cells.
Estrogen receptor alpha (ERα) and estrogen receptor beta
(ERβ) associate with IFN-γ and this association modulates malignant
behaviour (53). In addition, PDIA1
interacts with ERα and regulates its structure and activity
(54). Therefore the difference of
the redox state between MCF-7 and MDA-MB-231 cells treated with
IFN-γ could be attributed to the fact that MCF-7 cells are
ERα and ERβ positive whereas MDA-MB-231 cells are triple
negative breast cancer cells. PDIA1 functions as ERα coregulator
modulating the receptors transcriptional activity (54). ERα modulates the gene expression of
growth hormone (GH) (55),
which alters cellular oxidative stress levels (56). The differential regulation of the
PDIA1-mediated GH gene expression in the ERα-positive MCF-7
cells and the ERα-negative MDA-MB-231 cells is an additional
potential explanation justifying the differential redox state in
the ERα positive vs. the ERα negative breast cancer cells.
ETOP treatment, on the other hand, inhibited ROS
generation in MCF-7 cells and induced oxidative stress in
MDA-MB-231 cells in the absence of PDIA1 suggesting that
this topoisomerase II inhibitor exerts its effects through distinct
pathways in the two cell lines. ETOP induces stabilization of the
tumor suppressor p53 and PDI modifies the activity of this tumor
suppressor (7) providing a
potential explanation for the observed differences in ROS
generation between the wild-type p53 expressing MCF-7 cells and the
MDA-MB-231 cells which bear mutated p53 (p53-R280K). The gene
expression of the NADPH oxidase family member Nox4, which is
a source of ROS, has been demonstrated to be differentially
regulated by wt and mutant p53 (57). Evidence indicating association
between PDI and p53 (7) as well as
PDI and Nox4 (58) has been
previously presented suggesting that differences in the ROS levels
generated in the MCF-7 compared to MDA-MB-231 could be attributed
to the differential Nox4 levels in the two cell lines.
Glutathione is a known modulator of the function of
PDIA1 (59) regulating the ratio of
the oxidized vs. the reduced PDIA1 conformations (60). Oxidized and reduced PDIA1
conformations or death outcome during ER stress (9) suggest the importance of the interplay
between GSH and PDIA1 in cancer progression. Silencing of
PDIA1 increased cellular GSH concentration in MCF-7 cells
whereas the opposite was the case in MDA-MB-231 cells where in the
absence of PDIA1 decreased GSH levels were observed. Since
depletion of cellular GSH is an indicator of apoptosis initiation
(61), the obtained results
indicate that PDIA1 is a pro-apoptotic factor in MCF-7 cells
whereas in MDA-MB-231 cells it plays a pro-survival role. This
conclusion is supported by the results obtained from the
experiments assessing MOMP demonstrating that silencing of
PDIA1 reduced the polarization of the mitochondrial membrane
in untreated MCF-7 cells and did not affect mitochondrial membrane
disruption in the MDA-MB-231 cells under the same conditions. A
potential mechanism justifying the differential effects of
PDIA1 silencing on mitochondrial membrane potential in the
two cell lines is the mitochondrial colocalization of the PDIA1
(13) with the estrogen receptors
ERα and ERβ in the MCF-7 breast cancer cells (62). Mitochondrial localization of ERα is
associated with the modulation of the mitochondrial membrane
potential and the inhibition of mitochondrial ROS generation due to
the upregulation of the manganese superoxide dismutase activity in
MCF-7 ERα-positive cells (62).
The observed changes in the ROS generation,
regulation of antioxidant cellular levels and mitochondrial
membrane potential in the presence vs. the absence of PDIA1
are indications that PDIA1 plays a critical role in the
communication between endoplasmic reticulum and mitochondria and as
such in the regulation of mitochondrial biogenesis and potentially
energy metabolism (63). Support to
this hypothesis is lent by observations showing the relationship
between calcium and energy metabolism and the link between PDIA1
and the regulation of calcium homeostasis (64). Measurement of ATP production
indicated that MCF-7 cells produced higher ATP levels in the
absence of PDIA1 whereas MDA-MB-231 in the absence of
PDIA1 produced significantly lower ATP levels. The crosstalk
between PDIA1, p53 and Nox4 in the regulation of the activity of
the mitochondrial respiratory chain (65,66)
and thus ATP production may be the reason for the differential ATP
levels observed in the MCF-7 and MDA-MB-231 cells.
The immune system recognizes and eliminates
neoplastic cells by identifying tumor specific antigens presented
to the immune system cells in complex with MHC class I molecules.
The cellular redox state is a crucial factor contributing to the
efficient recognition of tumor antigens by the immune system
(25) and PDIA1 governs the antigen
processing and presentation events (22). Apart from its role in the
stabilization of the early MHC class I complex and selection of the
appropriate antigen PDIA1 has also been shown to participate in the
regulation of the expression of the MHC class I (46). The expression of the classical or
non-classical type of MHC class I is a critical point
distinguishing the visible from the invisible to the immune system
tumors, as overexpression of HLA-G facilitates evasion of
the immune surveillance by the tumor cells (50,67).
HLA-G has been shown to inactivate the effector function of the
natural killer (NK) cells by associating with the inhibitory
receptor of these cells (51).
Results shown in Fig. 6 indicate
that the HLA-G cell surface levels in the cells expressing
PDIA1 are higher compared to those in cells in which the
expression of PDIA1 had been silenced implying that by
regulating the HLA-G surface levels PDIA1 potentially
facilitates tumor cells to escape NK cell mediated innate immune
responses thereby promoting immunotolerance (52).
PDIA1 overexpression has been reported in
several types of cancer and is correlated with metastasis and
resistance to cancer therapy (68,69).
PDIA1 has also been shown to associate with well-characterized
metastatic factors including metalloproteases, selectins and
integrins (68,70). Results shown in this study
indicating correlation between the PDIA1/HLA-G mRNA ratio
and overall survival in breast cancer patients provides further
support for the hypothesis that PDIA1 is involved in the
co-ordination of immune responses to tumor cells as well as in
metastasis. Furthermore, the fact that high PDIA1 and low
HLA-G mRNA ratio was found in the subgroup of the stage 2
ERα-positive breast cancer patients exhibiting low PDIA1 and high
HLA-G mRNA levels the longer they survive, whereas low PDIA1
and high HLA-G mRNA ratio was measured in the longer
survivors of the same subgroup of ERα-negative breast cancer
patients could provide the potential means for selective treatment
of the two different types of patients.
PDIA1 plays differential role in the regulation of
the cellular redox state in the ERα-positive MCF-7 vs. the TNBC
MDA-MB-231 cells. In particular, silencing of PDIA1
downregulated ROS levels in MCF-7 cells and upregulated ROS levels
in MDA-MB-231 cells. Upregulation of GSH levels in PDIA1
silenced MCF-7 cells and downregulation of GSH levels in
PDIA1 silenced MDA-MB-231 cells suggesting that PDIA1 is a
pro-apoptotic factor in the former and pro-survival in the latter
cells. ATP production was not affected in MCF-7 cells whereas
MDA-MB-231 cells in which PDIA1 had been silenced produced
lower ATP levels compared to PDIA1 expressing cells. The positive
correlation of PDIA1 mRNA levels with HLA-G gene
expression in breast cancer patients together with results showing
downregulation of HLA-G levels on the extracellular membrane of
MCF-7 and MDA-MB-231 cells lacking PDIA1 suggest that PDIA1
may contribute to the evasion of the immune surveillance by breast
cancer cells. In addition the correlation of the ratio of PDIA1
and HLA-G mRNA levels in stage 2 breast cancer patients
indicates that PDIA1 could be used as a determining factor in the
stratification of patients that would be responsive to
immunotherapy.
Supplementary Material
Supporting Data
Acknowledgements
Part of this study was included in R. Alhammads PhD
thesis. We would like to thank Gruppo Italiano Mesotelioma for
support.
Funding
The study received funding from the Staff
Development Fund, Naresuan University, Thailand Research Fund-Royal
Golden Jubilee Ph.D. Program of Thailand and the British Council
Newton Fund, the Center of Excellence on Medical Biotechnology
(CEMB) (grant no. CEMB-RP-005), the Thailand Research Fund
International Research Network (TRF-IRN) (grant no. IRN58W001), and
the Siriraj Research Fund (grant no. R016034008), and the Siriraj
Chalermphrakiat Grant.
Availability of data and materials
All data generated or analysed during this study
are included in this published article
Authors contributions
RA planned and performed experiments, analysed the
results and prepared the draft of the manuscript, SK and NT
performed experiments, analysed the results and reviewed the
manuscript. TL, PY and LM analysed the results and reviewed the
manuscript. MKD and CD have formed the hypothesis and supervised
the research carried out, interpreted the results and prepared the
manuscript. All authors approved the final manuscript.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PDI
|
protein disulphide isomerase
|
|
ROS
|
reactive oxygen species
|
|
TNBC
|
triple negative breast cancer
|
|
MMD
|
mitochondrial membrane disruption
|
|
RISC
|
RNA-interfering silencing complex
|
References
|
1
|
Liou GY and Storz P: Reactive oxygen
species in cancer. Free Radic Res. 44:479–496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schieber M and Chandel NS: ROS function in
redox signaling and oxidative stress. Curr Biol. 24:R453–R462.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eletto D, Chevet E, Argon Y and
Appenzeller-Herzog C: Redox controls UPR to control redox. J Cell
Sc. 127:3649–3658. 2014. View Article : Google Scholar
|
|
4
|
Almanza A, Carlesso A, Chintha C,
Creedican S, Doultsinos D, Leuzzi B, Luis A, McCarthy N,
Montibeller L, More S, et al: Endoplasmic reticulum stress
signalling-from basic mechanisms to clinical applications. FEBS J.
286:241–278. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oakes SA: Endoplasmic reticulum
proteostasis: A key checkpoint in cancer. Am J Physiol Cell
Physiol. 312:C93–C102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Z, Zhang L, Zhou L, Lei Y, Zhang Y
and Huang C: Redox signaling and unfolded protein response
coordinate cell fate decisions under ER stress. Redox Biol.
25:1010472018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kranz P, Neumann F, Wolf A, Classen F,
Pompsch M, Ocklenburg T, Baumann J, Janke K, Baumann M, Goepelt K,
et al: PDI is an essential redox-sensitive activator of PERK during
the unfolded protein response (UPR). Cell Death Dis. 8:e29862017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okada K, Fukui M and Zhu BT: Protein
disulfide isomerase mediates glutathione depletion-induced
cytotoxicity. Biochem Biophys Res Commun. 477:495–502. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grek C and Townsend DM: Protein disulfide
isomerase superfamily in disease and the regulation of apoptosis.
Endoplasmic Reticulum Stress Dis. 1:4–17. 2014.PubMed/NCBI
|
|
10
|
Ali Khan H and Mutus B: Protein disulfide
isomerase a multifunctional protein with multiple physiological
roles. Front Chem. 2:702014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang C, Li W, Ren J, Fang J, Ke H, Gong W,
Feng W and Wang CC: Structural insights into the redox-regulated
dynamic conformations of human protein disulfide isomerase.
Antioxid Redox Signal. 19:36–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu J, Li T, Liu Y, Wang X, Zhang J, Shi G,
Lou J and Wang L, Wang CC and Wang L: Phosphorylation switches
protein disulfide isomerase activity to maintain proteostasis and
attenuate ER stress. EMBO J. 39:e1038412020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Turano C, Coppari S, Altieri F and Ferraro
A: Proteins of the PDI family: Unpredicted non-ER locations and
functions. J Cell Physiol. 193:154–163. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Parakh S and Atkin JD: Novel roles for
protein disulphide isomerase in disease states: A double edged
sword? Front Cell Dev Biol. 3:302015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao G, Lu H and Li C: Proapoptotic
activities of protein disulfide isomerase (PDI) and PDIA3 protein,
a role of the Bcl-2 protein Bak. J Biol Chem. 290:8949–8963. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ghanbari Movahed Z, Rastegari-Pouyani M,
Mohammadi MH and Mansouri K: Cancer cells change their glucose
metabolism to overcome increased ROS: One step from cancer cell to
cancer stem cell? Biomed Pharmacother. 112:1086902019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Higuchi T, Watanabe Y and Waga I: Protein
disulfide isomerase suppresses the transcriptional activity of
NF-kappaB. Biochem Biophys Res Commun. 318:46–52. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goplen D, Wang J, Enger PO, Tysnes BB,
Terzis AJ, Laerum OD and Bjerkvig R: Protein disulfide isomerase
expression is related to the invasive properties of malignant
glioma. Cancer Res. 66:9895–9902. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reverendo M, Mendes A, Arguello RJ, Gatti
E and Pierre P: At the crossway of ER-stress and proinflammatory
responses. FEBS J. 286:297–310. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Obacz J, Avril T, Rubio-Patino C,
Bossowski JP, Igbaria A, Ricci JE and Chevet E: Regulation of
tumor-stroma interactions by the unfolded protein response. FEBS J.
286:279–296. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lamers M, Berlin I and Neefjes J: Antigen
presentation: Visualizing the MHC Class I peptide-loading
bottleneck. Curr Biol. 28:R83–R86. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang K, Park B, Oh C, Cho K and Ahn K: A
role for protein disulfide isomerase in the early folding and
assembly of MHC class I molecules. Antioxid Redox Signal.
11:2553–2561. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park B, Lee S, Kim E, Cho K, Riddell SR,
Cho S and Ahn K: Redox regulation facilitates optimal peptide
selection by MHC class I during antigen processing. Cell.
127:369–382. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cho K, Cho S, Lee SO, Oh C, Kang K, Ryoo
J, Lee S, Kang S and Ahn K: Redox-regulated peptide transfer from
the transporter associated with antigen processing to major
histocompatibility complex class I molecules by protein disulfide
isomerase. Antioxid Redox Signal. 15:621–633. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee S, Park B, Kang K and Ahn K:
Redox-regulated export of the major histocompatibility complex
class I-peptide complexes from the endoplasmic reticulum. Mol Biol
Cell. 20:3285–3294. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Soares Moretti AI and Martins Laurindo FR:
Protein disulfide isomerases: Redox connections in and out of the
endoplasmic reticulum. Arch Biochem Biophys. 617:106–119. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guazzelli A, Meysami P, Bakker E,
Demonacos C, Giordano A, Krstic-Demonacos M and Mutti L: BAP1
status determines the sensitivity of malignant mesothelioma cells
to gemcitabine treatment. Int J Mol Sci. 20:4292019. View Article : Google Scholar
|
|
29
|
Forkink M, Smeitink JA, Brock R, Willems
PH and Koopman WJ: Detection and manipulation of mitochondrial
reactive oxygen species in mammalian cells. Biochim Biophys Acta.
1797:1034–1044. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cereda M, Gambardella G, Benedetti L,
Iannelli F, Patel D, Basso G, Guerra RF, Mourikis TP, Puccio I,
Sinha S, et al: Patients with genetically heterogeneous synchronous
colorectal cancer carry rare damaging germline mutations in
immune-related genes. Nat Commun. 7:120722016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barrey E, Saint-Auret G, Bonnamy B, Damas
D, Boyer O and Gidrol X: Pre-microRNA and mature microRNA in human
mitochondria. PLoS One. 6:e202202011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zibara K, Zeidan A, Bjeije H, Kassem N,
Badran B and El-Zein N: ROS mediates interferon gamma induced
phosphorylation of Src, through the Raf/ERK pathway, in MCF-7 human
breast cancer cell line. J Cell Commun Signal. 11:57–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oh SY, Sohn YW, Park JW, Park HJ, Jeon HM,
Kim TK, Lee JS, Jung JE, Jin X, Chung YG, et al: Selective cell
death of oncogenic Akt-transduced brain cancer cells by etoposide
through reactive oxygen species mediated damage. Mol Cancer Ther.
6:2178–2187. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hodny Z, Reinis M, Hubackova S, Vasicova P
and Bartek J: Interferon gamma/NADPH oxidase defense system in
immunity and cancer. Oncoimmunology. 5:e10804162016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Laurindo FR, Pescatore LA and Fernandes
Dde C: Protein disulfide isomerase in redox cell signaling and
homeostasis. Free Radic Biol Med. 52:1954–1969. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hegedűs C, Kovács K, Polgár Z, Regdon Z,
Szabó É, Robaszkiewicz A, Forman HJ, Martner A and Virág L: Redox
control of cancer cell destruction. Redox Biol. 16:59–74. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Traverso N, Ricciarelli R, Nitti M,
Marengo B, Furfaro AL, Pronzato MA, Marinari UM and Domenicotti C:
Role of glutathione in cancer progression and chemoresistance. Oxid
Med Cell Longev. 2013:9729132013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bulleid NJ: Disulfide bond formation in
the mammalian endoplasmic reticulum. Cold Spring Harb Perspect
Biol. 4:a0132192012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Herrera-Cruz MS and Simmen T: Cancer:
Untethering mitochondria from the endoplasmic reticulum? Front
Oncol. 7:1052017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Koczian F, Naglo O, Vomacka J, Vick B,
Servatius P, Zisis T, Hettich B, Kazmaier U, Sieber SA, Jeremias I,
et al: Targeting the endoplasmic reticulum-mitochondria interface
sensitizes leukemia cells to cytostatics. Haematologica.
104:546–555. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Marchi S, Patergnani S and Pinton P: The
endoplasmic reticulum-mitochondria connection: One touch, multiple
functions. Biochim Biophys Acta. 1837:461–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kerkhofs M, Bittremieux M, Morciano G,
Giorgi C, Pinton P, Parys JB and Bultynck G: Emerging molecular
mechanisms in chemotherapy: Ca(2+) signaling at the
mitochondria-associated endoplasmic reticulum membranes. Cell Death
Dis. 9:3342018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoboue ED, Sitia R and Simmen T: Redox
crosstalk at endoplasmic reticulum (ER) membrane contact sites
(MCS) uses toxic waste to deliver messages. Cell Death Dis.
9:3312018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wacquier B, Combettes L and Dupont G:
Cytoplasmic and mitochondrial calcium signaling: A two-way
relationship. Cold Spring Harb Perspect Biol. 11:a0351392019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
So JS: Roles of endoplasmic reticulum
stress in immune responses. Mol Cells. 41:705–716. 2018.PubMed/NCBI
|
|
46
|
Kukita K, Tamura Y, Tanaka T, Kajiwara T,
Kutomi G, Saito K, Okuya K, Takaya A, Kanaseki T, Tsukahara T, et
al: Cancer-associated oxidase ERO1-α regulates the expression of
MHC class I molecule via oxidative folding. J Immunol.
194:4988–4996. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim Y, Kang K, Kim I, Lee YJ, Oh C, Ryoo
J, Jeong E and Ahn K: Molecular mechanisms of MHC class I-antigen
processing: Redox considerations. Antioxid Redox Signal.
11:907–936. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Binder RJ: Functions of heat shock
proteins in pathways of the innate and adaptive immune system. J
Immunol. 193:5765–5771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Allen RL: Non-classical immunology. Genome
Biol. 2:REPORTS40042001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
da Silva GB, Silva TG, Duarte RA, Neto NL,
Carrara HH, Donadi EA, Goncalves MA, Soares EG and Soares CP:
Expression of the classical and nonclassical HLA molecules in
breast cancer. Int J Breast Cancer. 2013:2504352013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sivori S, Vacca P, Del Zotto G, Munari E,
Mingari MC and Moretta L: Human NK cells: Surface receptors,
inhibitory checkpoints, and translational applications. Cell Mol
Immunol. 16:430–441. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Morandi F, Rizzo R, Fainardi E,
Rouas-Freiss N and Pistoia V: Recent advances in our understanding
of HLA-G biology: Lessons from a wide spectrum of human diseases. J
Immunol Res. 2016:43264952016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Niu XL, Wang Y, Yao Z, Duan H, Li Z, Liu
W, Zhang H and Deng WM: Autocrine interferon-gamma may affect
malignant behavior and sensitivity to tamoxifen of MCF-7 via
estrogen receptor β subtype. Oncol Rep. 34:3120–3130. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schultz-Norton JR, McDonald WH, Yates JR
and Nardulli AM: Protein disulfide isomerase serves as a molecular
chaperone to maintain estrogen receptor alpha structure and
function. Mol Endocrinol. 20:1982–1995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hashimoto S and Imaoka S:
Protein-disulfide isomerase regulates the thyroid hormone
receptor-mediated gene expression via redox factor-1 through thiol
reduction-oxidation. J Biol Chem. 288:1706–1716. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mancini A, Bruno C, Vergani E, Guidi F,
Angelini F, Meucci E and Silvestrini A: Evaluation of oxidative
stress effects on different macromolecules in adult growth hormone
deficiency. PLoS One. 15:e02363572020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boudreau HE, Casterline BW, Burke DJ and
Leto TL: Wild-type and mutant p53 differentially regulate NADPH
oxidase 4 in TGF-β-mediated migration of human lung and breast
epithelial cells. Br J Cancer. 110:2569–2582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Trevelin SC and Lopes LR: Protein
disulfide isomerase and Nox: New partners in redox signaling. Curr
Pharm Des. 21:5951–5963. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lappi AK and Ruddock LW: Reexamination of
the role of interplay between glutathione and protein disulfide
isomerase. J Mol Biol. 409:238–249. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hudson DA, Gannon SA and Thorpe C:
Oxidative protein folding: From thiol-disulfide exchange reactions
to the redox poise of the endoplasmic reticulum. Free Radic Biol
Med. 80:171–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Coppola S and Ghibelli L: GSH extrusion
and and the mitochondrial pathway of apoptotic signalling. Biochem
Soc Trans. 28:56–61. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pedram A, Razandi M, Wallace DC and Levin
ER: Functional estrogen receptors in the mitochondria of breast
cancer cells. Mol Biol Cell. 17:2125–2137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fan Y and Simmen T: Mechanistic
connections between endoplasmic reticulum (ER) redox control and
mitochondrial metabolism. Cells. 8:10712019. View Article : Google Scholar
|
|
64
|
Gutierrez T and Simmen T: Endoplasmic
reticulum chaperones tweak the mitochondrial calcium rheostat to
control metabolism and cell death. Cell Calcium. 70:64–75. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dikalov S: Cross talk between mitochondria
and NADPH oxidases. Free Radic Biol Med. 51:1289–1301. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kozieł R, Pircher H, Kratochwil M, Lener
B, Hermann M, Dencher NA and Jansen-Dürr P: Mitochondrial
respiratory chain complex I is inactivated by NADPH oxidase Nox4.
Biochem J. 452:231–239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kochan G, Escors D, Breckpot K and
Guerrero-Setas D: Role of non-classical MHC class I molecules in
cancer immunosuppression. Oncoimmunology. 2:e264912013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xu S, Sankar S and Neamati N: Protein
disulfide isomerase: A promising target for cancer therapy. Drug
Discov Today. 19:222–240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lee E and Lee DH: Emerging roles of
protein disulfide isomerase in cancer. BMB Rep. 50:401–410. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rosenberg N, Mor-Cohen R, Sheptovitsky VH,
Romanenco O, Hess O and Lahav J: Integrin-mediated cell adhesion
requires extracellular disulfide exchange regulated by protein
disulfide isomerase. Exp Cell Res. 381:77–85. 2019. View Article : Google Scholar : PubMed/NCBI
|